Characterization of the ATPase activity of RecG and RuvAB proteins on model fork structures reveals insight into stalled DNA replication fork repair
- PMID: 23893472
- PMCID: PMC3772186
- DOI: 10.1074/jbc.M113.500223
Characterization of the ATPase activity of RecG and RuvAB proteins on model fork structures reveals insight into stalled DNA replication fork repair
Abstract
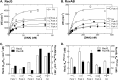
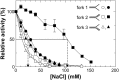
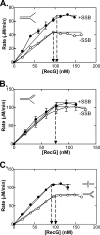
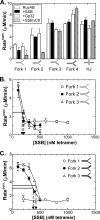
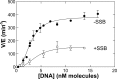
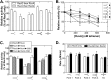
RecG and RuvAB are proposed to act at stalled DNA replication forks to facilitate replication restart. To clarify the roles of these proteins in fork regression, we used a coupled spectrophotometric ATPase assay to determine how these helicases act on two groups of model fork substrates: the first group mimics nascent stalled forks, whereas the second mimics regressed fork structures. The results show that RecG is active on the substrates in group 1, whereas these are poor substrates for RuvAB. In addition, in the presence of group 1 forks, the single-stranded DNA-binding protein (SSB) enhances the activity of RecG and enables it to compete with excess RuvA. In contrast, SSB inhibits the activity of RuvAB on these substrates. Results also show that the preferred regressed fork substrate for RuvAB is a Holliday junction, not a forked DNA. The active form of the enzyme on the Holliday junction contains a single RuvA tetramer. In contrast, although the enzyme is active on a regressed fork structure, RuvB loading by a single RuvA tetramer is impaired, and full activity requires the cooperative binding of two forked DNA substrate molecules. Collectively, the data support a model where RecG is responsible for stalled DNA replication fork regression. SSB ensures that if the nascent fork has single-stranded DNA character RuvAB is inhibited, whereas the activity of RecG is preferentially enhanced. Only once the fork has been regressed and the DNA is relaxed can RuvAB bind to a RecG-extruded Holliday junction.
Keywords: ATPases; DNA Recombination; DNA-Protein Interaction; DNA-binding Protein; Enzyme Kinetics; Fork Regression; Holliday Junction; RecG; Replication Fork; RuvAB.
Figures

References
-
- Kowalczykowski S. C. (2000) Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25, 156–165 - PubMed
-
- Seigneur M., Bidnenko V., Ehrlich S. D., Michel B. (1998) RuvAB acts at arrested replication forks. Cell 95, 419–430 - PubMed
-
- Cox M. M., Goodman M. F., Kreuzer K. N., Sherratt D. J., Sandler S. J., Marians K. J. (2000) The importance of repairing stalled replication forks. Nature 404, 37–41 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

