A microfluidic device for preparing next generation DNA sequencing libraries and for automating other laboratory protocols that require one or more column chromatography steps
- PMID: 23894273
- PMCID: PMC3722208
- DOI: 10.1371/journal.pone.0064084
A microfluidic device for preparing next generation DNA sequencing libraries and for automating other laboratory protocols that require one or more column chromatography steps
Abstract
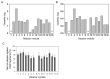
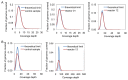
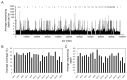
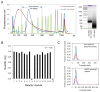
Library preparation for next-generation DNA sequencing (NGS) remains a key bottleneck in the sequencing process which can be relieved through improved automation and miniaturization. We describe a microfluidic device for automating laboratory protocols that require one or more column chromatography steps and demonstrate its utility for preparing Next Generation sequencing libraries for the Illumina and Ion Torrent platforms. Sixteen different libraries can be generated simultaneously with significantly reduced reagent cost and hands-on time compared to manual library preparation. Using an appropriate column matrix and buffers, size selection can be performed on-chip following end-repair, dA tailing, and linker ligation, so that the libraries eluted from the chip are ready for sequencing. The core architecture of the device ensures uniform, reproducible column packing without user supervision and accommodates multiple routine protocol steps in any sequence, such as reagent mixing and incubation; column packing, loading, washing, elution, and regeneration; capture of eluted material for use as a substrate in a later step of the protocol; and removal of one column matrix so that two or more column matrices with different functional properties can be used in the same protocol. The microfluidic device is mounted on a plastic carrier so that reagents and products can be aliquoted and recovered using standard pipettors and liquid handling robots. The carrier-mounted device is operated using a benchtop controller that seals and operates the device with programmable temperature control, eliminating any requirement for the user to manually attach tubing or connectors. In addition to NGS library preparation, the device and controller are suitable for automating other time-consuming and error-prone laboratory protocols requiring column chromatography steps, such as chromatin immunoprecipitation.
Conflict of interest statement
Figures

References
-
- Mitchell P (2001) Microfluidics--downsizing large-scale biology. Nat Biotechnol 19: 717-721. - PubMed
-
- Hong JW, Studer V, Hang G, Anderson WF, Quake SR (2004) A nanoliter-scale nucleic acid processor with parallel architecture. Nat Biotechnol 22: 435-439. - PubMed
-
- Marcus JS, Anderson WF, Quake SR (2006) Microfluidic single-cell mRNA isolation and analysis. Anal Chem 78: 3084-3089 - PubMed
-
- Hong JW, Chen Y, Anderson WF, Quake SR (2006) Molecular biology on a microfluidic chip. J Phys Condens Matter 18: S691.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

