Culture-independent investigation of the microbiome associated with the nematode Acrobeloides maximus
- PMID: 23894287
- PMCID: PMC3718782
- DOI: 10.1371/journal.pone.0067425
Culture-independent investigation of the microbiome associated with the nematode Acrobeloides maximus
Abstract
Background: Symbioses between metazoans and microbes are widespread and vital to many ecosystems. Recent work with several nematode species has suggested that strong associations with microbial symbionts may also be common among members of this phylu. In this work we explore possible symbiosis between bacteria and the free living soil bacteriovorous nematode Acrobeloides maximus.
Methodology: We used a soil microcosm approach to expose A. maximus populations grown monoxenically on RFP labeled Escherichia coli in a soil slurry. Worms were recovered by density gradient separation and examined using both culture-independent and isolation methods. A 16S rRNA gene survey of the worm-associated bacteria was compared to the soil and to a similar analysis using Caenorhabditis elegans N2. Recovered A. maximus populations were maintained on cholesterol agar and sampled to examine the population dynamics of the microbiome.
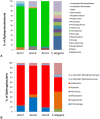
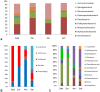
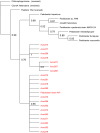
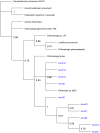
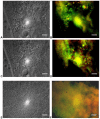
Results: A consistent core microbiome was extracted from A. maximus that differed from those in the bulk soil or the C. elegans associated set. Three genera, Ochrobactrum, Pedobacter, and Chitinophaga, were identified at high levels only in the A. maximus populations, which were less diverse than the assemblage associated with C. elegans. Putative symbiont populations were maintained for at least 4 months post inoculation, although the levels decreased as the culture aged. Fluorescence in situ hybridization (FISH) using probes specific for Ochrobactrum and Pedobacter stained bacterial cells in formaldehyde fixed nematode guts.
Conclusions: Three microorganisms were repeatedly observed in association with Acrobeloides maximus when recovered from soil microcosms. We isolated several Ochrobactrum sp. and Pedobacter sp., and demonstrated that they inhabit the nematode gut by FISH. Although their role in A. maximus is not resolved, we propose possible mutualistic roles for these bacteria in protection of the host against pathogens and facilitating enzymatic digestion of other ingested bacteria.
Conflict of interest statement
Figures


References
-
- Landmann F, Voronin D, Sullivan W, Taylor MJ (2011) Anti-filarial Activity of Antibiotic Therapy Is Due to Extensive Apoptosis after Wolbachia Depletion from Filarial Nematodes. PLOS Pathogens 7 (11) e1002351 doi:10.1371/journal.ppat.1002351 - DOI - PMC - PubMed
-
- An R, Grewal PS (2010) Molecular mechanisms of persistence of mutualistic bacteria Photorhabdus in the entomopathogenic nematode host. PLoS One 5 (10) e13154 doi:10.1371/journal.pone.0013154 - DOI - PMC - PubMed
-
- Clarke DJ (2008) Photorhabdus: a model for the analysis of pathogenicity and mutualism. Cellular Microbiology 10: 2159–2167. - PubMed
-
- Duchaud E, Rusniok C, Frangeul L, Buchrieser C, Givaudan A, et al. (2003) The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nature Biotechnology 21: 1307–1313. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

