Complete genome sequence of the cystic fibrosis pathogen Achromobacter xylosoxidans NH44784-1996 complies with important pathogenic phenotypes
- PMID: 23894309
- PMCID: PMC3718787
- DOI: 10.1371/journal.pone.0068484
Complete genome sequence of the cystic fibrosis pathogen Achromobacter xylosoxidans NH44784-1996 complies with important pathogenic phenotypes
Abstract
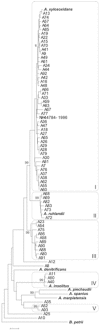
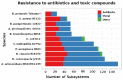
Achromobacter xylosoxidans is an environmental opportunistic pathogen, which infects an increasing number of immunocompromised patients. In this study we combined genomic analysis of a clinical isolated A. xylosoxidans strain with phenotypic investigations of its important pathogenic features. We present a complete assembly of the genome of A. xylosoxidans NH44784-1996, an isolate from a cystic fibrosis patient obtained in 1996. The genome of A. xylosoxidans NH44784-1996 contains approximately 7 million base pairs with 6390 potential protein-coding sequences. We identified several features that render it an opportunistic human pathogen, We found genes involved in anaerobic growth and the pgaABCD operon encoding the biofilm adhesin poly-β-1,6-N-acetyl-D-glucosamin. Furthermore, the genome contains a range of antibiotic resistance genes coding efflux pump systems and antibiotic modifying enzymes. In vitro studies of A. xylosoxidans NH44784-1996 confirmed the genomic evidence for its ability to form biofilms, anaerobic growth via denitrification, and resistance to a broad range of antibiotics. Our investigation enables further studies of the functionality of important identified genes contributing to the pathogenicity of A. xylosoxidans and thereby improves our understanding and ability to treat this emerging pathogen.
Conflict of interest statement
Figures



References
-
- De Baets F, Schelstraete P, Van Daele S, Haerynck F, Vaneechoutte M (2007) Achromobacter xylosoxidans in cystic fibrosis: prevalence and clinical relevance. J Cyst Fibros 6: 75-78. doi:10.1016/j.jcf.2006.05.011. PubMed: 16793350. - DOI - PubMed
-
- Gómez-Cerezo J, Suárez I, Ríos JJ, Peña P, García de Miguel MJ et al. (2003) Achromobacter xylosoxidans bacteremia: a 10-year analysis of 54 cases. Eur J Clin Microbiol Infect Dis 22: 360-363. doi:10.1007/s10096-003-0925-3. PubMed: 12750959. - DOI - PubMed
-
- Klinger JD, Thomassen MJ (1985) Occurrence and antimicrobial susceptibility of gram-negative nonfermentative bacilli in cystic fibrosis patients. Diagn Microbiol Infect Dis 3: 149-158. doi:10.1016/0732-8893(85)90025-2. PubMed: 3979021. - DOI - PubMed
-
- Liu L, Coenye T, Burns JL, Whitby PW, Stull TL et al. (2002) Ribosomal DNA-directed PCR for identification of Achromobacter (Alcaligenes) xylosoxidans recovered from sputum samples from cystic fibrosis patients. J Clin Microbiol 40: 1210-1213. doi:10.1128/JCM.40.4.1210-1213.2002. PubMed: 11923333. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

