Lactobacillus paracasei comparative genomics: towards species pan-genome definition and exploitation of diversity
- PMID: 23894338
- PMCID: PMC3716772
- DOI: 10.1371/journal.pone.0068731
Lactobacillus paracasei comparative genomics: towards species pan-genome definition and exploitation of diversity
Abstract
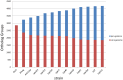
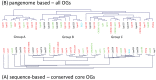
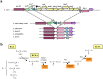
Lactobacillus paracasei is a member of the normal human and animal gut microbiota and is used extensively in the food industry in starter cultures for dairy products or as probiotics. With the development of low-cost, high-throughput sequencing techniques it has become feasible to sequence many different strains of one species and to determine its "pan-genome". We have sequenced the genomes of 34 different L. paracasei strains, and performed a comparative genomics analysis. We analysed genome synteny and content, focussing on the pan-genome, core genome and variable genome. Each genome was shown to contain around 2800-3100 protein-coding genes, and comparative analysis identified over 4200 ortholog groups that comprise the pan-genome of this species, of which about 1800 ortholog groups make up the conserved core. Several factors previously associated with host-microbe interactions such as pili, cell-envelope proteinase, hydrolases p40 and p75 or the capacity to produce short branched-chain fatty acids (bkd operon) are part of the L. paracasei core genome present in all analysed strains. The variome consists mainly of hypothetical proteins, phages, plasmids, transposon/conjugative elements, and known functions such as sugar metabolism, cell-surface proteins, transporters, CRISPR-associated proteins, and EPS biosynthesis proteins. An enormous variety and variability of sugar utilization gene cassettes were identified, with each strain harbouring between 25-53 cassettes, reflecting the high adaptability of L. paracasei to different niches. A phylogenomic tree was constructed based on total genome contents, and together with an analysis of horizontal gene transfer events we conclude that evolution of these L. paracasei strains is complex and not always related to niche adaptation. The results of this genome content comparison was used, together with high-throughput growth experiments on various carbohydrates, to perform gene-trait matching analysis, in order to link the distribution pattern of a specific phenotype to the presence/absence of specific sets of genes.
Conflict of interest statement
Figures



References
-
- Salminen S, von Wright A, Ouwehand AC (2004) Lactic Acid Bacteria: Microbiological and Functional Aspects. New York: Marcel Dekker, Inc.
-
- Holzapfel WH, Wood BJB (1998) The genera of lactic acid bacteria London Blackie Academic & Professional.
-
- de Vrese M, Schrezenmeir J (2008) Probiotics, prebiotics, and synbiotics. Adv Biochem Eng Biotechnol 111: 1–66. - PubMed
-
- Marchand J, Vandenplas Y (2000) Micro-organisms administered in the benefit of the host: myths and facts. Eur J Gastroenterol Hepatol 12: 1077–1088. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

