Epigenetic regulation of cardiac progenitor cells marker c-kit by stromal cell derived factor-1α
- PMID: 23894420
- PMCID: PMC3722185
- DOI: 10.1371/journal.pone.0069134
Epigenetic regulation of cardiac progenitor cells marker c-kit by stromal cell derived factor-1α
Retraction in
-
Retraction: Epigenetic Regulation of Cardiac Progenitor Cells Marker c-kit by Stromal Cell Derived Factor-1α.PLoS One. 2021 Feb 12;16(2):e0247094. doi: 10.1371/journal.pone.0247094. eCollection 2021. PLoS One. 2021. PMID: 33577587 Free PMC article. No abstract available.
Abstract
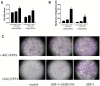
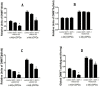
Background: Cardiac progenitor cells (CPCs) have been proven suitable for stem cell therapy after myocardial infarction, especially c-kit(+)CPCs. CPCs marker c-kit and its ligand, the stem cell factor (SCF), are linked as c-kit/SCF axis, which is associated with the functions of proliferation and differentiation. In our previous study, we found that stromal cell-derived factor-1α (SDF-1α) could enhance the expression of c-kit. However, the mechanism is unknown.
Methods and results: CPCs were isolated from adult mouse hearts, c-kit(+) and c-kit(-) CPCs were separated by magnetic beads. The cells were cultured with SDF-1α and CXCR4-selective antagonist AMD3100, and c-kit expression was measured by qPCR and Western blotting. Results showed that SDF-1α could enhance c-kit expression of c-kit(+)CPCs, made c-kit(-)CPCs expressing c-kit, and AMD3100 could inhibit the function of SDF-1α. After the intervention of SDF-1α and AMD3100, proliferation and migration of CPCs were measured by CCK-8 and transwell assay. Results showed that SDF-1α could enhance the proliferation and migration of both c-kit(+) and c-kit(-) CPCs, and AMD3100 could inhibit these functions. DNA methyltransferase (DNMT) mRNA were measured by qPCR, DNMT activity was measured using the DNMT activity assay kit, and DNA methylation was analyzed using Sequenom's MassARRAY platform, after the CPCs were cultured with SDF-1α. The results showed that SDF-1α stimulation inhibited the expression of DNMT1 and DNMT3β, which are critical for the maintenance of regional DNA methylation. Global DNMT activity was also inhibited by SDF-1α. Lastly, SDF-1α treatment led to significant demethylation in both c-kit(+) and c-kit(-) CPCs.
Conclusions: SDF-1α combined with CXCR4 could up-regulate c-kit expression of c-kit(+)CPCs and make c-kit(-)CPCs expressing c-kit, which result in the CPCs proliferation and migration ability improvement, through the inhibition of DNMT1 and DNMT3β expression and global DNMT activity, as well as the subsequent demethylation of the c-kit gene.
Conflict of interest statement
Figures



Similar articles
-
Regulation of c-kit+ progenitor cells by stromal cell derived factor-1α in adult murine heart.Heart Lung Circ. 2014 Jan;23(1):75-81. doi: 10.1016/j.hlc.2013.05.652. Epub 2013 Jul 23. Heart Lung Circ. 2014. PMID: 23891309
-
Hypoxic preconditioning improves survival of cardiac progenitor cells: role of stromal cell derived factor-1α-CXCR4 axis.PLoS One. 2012;7(7):e37948. doi: 10.1371/journal.pone.0037948. Epub 2012 Jul 18. PLoS One. 2012. PMID: 22815687 Free PMC article.
-
Overexpression of SDF-1α enhanced migration and engraftment of cardiac stem cells and reduced infarcted size via CXCR4/PI3K pathway.PLoS One. 2012;7(9):e43922. doi: 10.1371/journal.pone.0043922. Epub 2012 Sep 11. PLoS One. 2012. PMID: 22984452 Free PMC article.
-
Genetically manipulated progenitor/stem cells restore function to the infarcted heart via the SDF-1α/CXCR4 signaling pathway.Prog Mol Biol Transl Sci. 2012;111:265-84. doi: 10.1016/B978-0-12-398459-3.00012-5. Prog Mol Biol Transl Sci. 2012. PMID: 22917235 Review.
-
After the storm: an objective appraisal of the efficacy of c-kit+ cardiac progenitor cells in preclinical models of heart disease.Can J Physiol Pharmacol. 2021 Feb;99(2):129-139. doi: 10.1139/cjpp-2020-0406. Epub 2020 Sep 16. Can J Physiol Pharmacol. 2021. PMID: 32937086 Free PMC article. Review.
Cited by
-
Insulin-like growth factor-1-mediated regulation of miR-193a expression promotes the migration and proliferation of c-kit-positive mouse cardiac stem cells.Stem Cell Res Ther. 2018 Feb 21;9(1):41. doi: 10.1186/s13287-017-0762-4. Stem Cell Res Ther. 2018. PMID: 29467020 Free PMC article.
-
Retraction: Epigenetic Regulation of Cardiac Progenitor Cells Marker c-kit by Stromal Cell Derived Factor-1α.PLoS One. 2021 Feb 12;16(2):e0247094. doi: 10.1371/journal.pone.0247094. eCollection 2021. PLoS One. 2021. PMID: 33577587 Free PMC article. No abstract available.
-
Multipotent stem cells of the heart-do they have therapeutic promise?Front Physiol. 2015 May 8;6:123. doi: 10.3389/fphys.2015.00123. eCollection 2015. Front Physiol. 2015. PMID: 26005421 Free PMC article. Review.
-
c-kit(+) cells: the tell-tale heart of cardiac regeneration?Cell Mol Life Sci. 2015 May;72(9):1725-40. doi: 10.1007/s00018-014-1832-8. Epub 2015 Jan 10. Cell Mol Life Sci. 2015. PMID: 25575564 Free PMC article. Review.
-
Age-Dependent Effect of Pediatric Cardiac Progenitor Cells After Juvenile Heart Failure.Stem Cells Transl Med. 2016 Jul;5(7):883-92. doi: 10.5966/sctm.2015-0241. Epub 2016 May 5. Stem Cells Transl Med. 2016. PMID: 27151913 Free PMC article.
References
-
- Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, et al. (2003) Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 114: 763–776. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

