Inactive DNMT3B splice variants modulate de novo DNA methylation
- PMID: 23894490
- PMCID: PMC3716610
- DOI: 10.1371/journal.pone.0069486
Inactive DNMT3B splice variants modulate de novo DNA methylation
Abstract
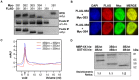
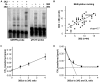
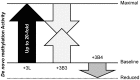
Inactive DNA methyltransferase (DNMT) 3B splice isoforms are associated with changes in DNA methylation, yet the mechanisms by which they act remain largely unknown. Using biochemical and cell culture assays, we show here that the inactive DNMT3B3 and DNMT3B4 isoforms bind to and regulate the activity of catalytically competent DNMT3A or DNMT3B molecules. DNMT3B3 modestly stimulated the de novo methylation activity of DNMT3A and also counteracted the stimulatory effects of DNMT3L, therefore leading to subtle and contrasting effects on activity. DNMT3B4, by contrast, significantly inhibited de novo DNA methylation by active DNMT3 molecules, most likely due to its ability to reduce the DNA binding affinity of co-complexes, thereby sequestering them away from their substrate. Immunocytochemistry experiments revealed that in addition to their effects on the intrinsic catalytic function of active DNMT3 enzymes, DNMT3B3 and DNMT34 drive distinct types of chromatin compaction and patterns of histone 3 lysine 9 tri-methylation (H3K9me3) deposition. Our findings suggest that regulation of active DNMT3 members through the formation of co-complexes with inactive DNMT3 variants is a general mechanism by which DNMT3 variants function. This may account for some of the changes in DNA methylation patterns observed during development and disease.
Conflict of interest statement
Figures



References
-
- Jurkowska RZ, Jurkowski TP, Jeltsch A (2011) Structure and function of mammalian DNA methyltransferases. Chembiochem 12: 206–222. - PubMed
-
- Goll MG, Bestor TH (2004) Eukaryotic Cytosine Methyltransferases. Annu Rev Biochem 74: 481–574. - PubMed
-
- Esteller M, Fraga MF, Paz MF, Campo E, Colomer D, et al... (2002) Cancer epigenetics and methylation. Science 297: 1807–1808; discussion 1807–1808. - PubMed
-
- Svedruzic ZM, Reich NO (2004) The mechanism of target base attack in DNA cytosine carbon 5 methylation. Biochemistry 43: 11460–11473. - PubMed
-
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, et al. (2007) The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450: 908–912. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

