PCR-induced transitions are the major source of error in cleaned ultra-deep pyrosequencing data
- PMID: 23894647
- PMCID: PMC3720931
- DOI: 10.1371/journal.pone.0070388
PCR-induced transitions are the major source of error in cleaned ultra-deep pyrosequencing data
Abstract
Background: Ultra-deep pyrosequencing (UDPS) is used to identify rare sequence variants. The sequence depth is influenced by several factors including the error frequency of PCR and UDPS. This study investigated the characteristics and source of errors in raw and cleaned UDPS data.
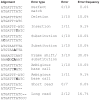
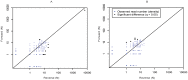
Results: UDPS of a 167-nucleotide fragment of the HIV-1 SG3Δenv plasmid was performed on the Roche/454 platform. The plasmid was diluted to one copy, PCR amplified and subjected to bidirectional UDPS on three occasions. The dataset consisted of 47,693 UDPS reads. Raw UDPS data had an average error frequency of 0.30% per nucleotide site. Most errors were insertions and deletions in homopolymeric regions. We used a cleaning strategy that removed almost all indel errors, but had little effect on substitution errors, which reduced the error frequency to 0.056% per nucleotide. In cleaned data the error frequency was similar in homopolymeric and non-homopolymeric regions, but varied considerably across sites. These site-specific error frequencies were moderately, but still significantly, correlated between runs (r=0.15-0.65) and between forward and reverse sequencing directions within runs (r=0.33-0.65). Furthermore, transition errors were 48-times more common than transversion errors (0.052% vs. 0.001%; p<0.0001). Collectively the results indicate that a considerable proportion of the sequencing errors that remained after data cleaning were generated during the PCR that preceded UDPS.
Conclusions: A majority of the sequencing errors that remained after data cleaning were introduced by PCR prior to sequencing, which means that they will be independent of platform used for next-generation sequencing. The transition vs. transversion error bias in cleaned UDPS data will influence the detection limits of rare mutations and sequence variants.
Conflict of interest statement
Figures


References
-
- Simen BB, Simons JF, Hullsiek KH, Novak RM, Macarthur RD, et al. (2009) Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis 199: 693–701. - PubMed
-
- Hirsch MS, Gunthard HF, Schapiro JM, Brun-Vezinet F, Clotet B, et al. (2008) Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin Infect Dis 47: 266–285. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

