Structural mechanisms underlying signaling in the cellular response to DNA double strand breaks
- PMID: 23896398
- PMCID: PMC3818410
- DOI: 10.1016/j.mrfmmm.2013.07.004
Structural mechanisms underlying signaling in the cellular response to DNA double strand breaks
Abstract
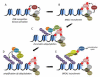
DNA double strand breaks (DSBs) constitute one of the most dangerous forms of DNA damage. In actively replicating cells, these breaks are first recognized by specialized proteins that initiate a signal transduction cascade that modulates the cell cycle and results in the repair of the breaks by homologous recombination (HR). Protein signaling in response to double strand breaks involves phosphorylation and ubiquitination of chromatin and a variety of associated proteins. Here we review the emerging structural principles that underlie how post-translational protein modifications control protein signaling that emanates from these DNA lesions.
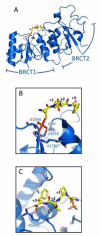
Keywords: BRCA1 C-terminal domain; BRCT; BRCT domains; DSB; Double-strand break signaling; FHA domains; FHAf; HR; MRN; Mre11–Rad50–NBS1; OTUB; Phosphorylation signaling; RIDDLE syndrome; UIM; Ubc13; Ubiquitin; double strand break; homologous recombination; orkhead-associated domain.; otubain or ovarian tumor domain protein; radiosensitivity, immunodeficiency, dysmorphic features and learning difficulties; ubiquitin interaction motif.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures


References
-
- Kasparek TR, Humphrey TC. DNA double-strand break repair pathways, chromosomal rearrangements and cancer. Semin Cell Dev Biol. 2011;22:886–897. - PubMed
-
- Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. - PubMed
-
- Caestecker KW, Van de Walle GR. The role of BRCA1 in DNA double-strand repair: past and present. Exp Cell Res. 2013;319:575–587. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

