AIEC pathobiont instigates chronic colitis in susceptible hosts by altering microbiota composition
- PMID: 23896971
- PMCID: PMC4089387
- DOI: 10.1136/gutjnl-2013-304909
AIEC pathobiont instigates chronic colitis in susceptible hosts by altering microbiota composition
Abstract
Background: Inflammatory bowel disease (IBD) is driven by a seemingly aberrant immune response to the gut microbiota with disease development dictated by genetics and environmental factors. A model exemplifying this notion is our recent demonstration that colonisation of adherent-invasive Escherichia coli (AIEC) during microbiota acquisition drove chronic colitis in mice lacking the flagellin receptor TLR5 (T5KO). T5KO colitis persisted beyond AIEC clearance and requires TLR4 and the NLRC4 inflammasome. We hypothesised that AIEC instigates chronic inflammation by increasing microbial lipopolysaccharide (LPS) and flagellin levels.
Goal: Examine if transient AIEC colonisation lastingly alters levels of LPS and flagellin and changes microbiota composition.
Methods: Germ-free mice (wild type (WT) and T5KO) were inoculated with AIEC strain LF82 and placed in standard housing allowing a complex microbiota that eliminated AIEC in both mice strains. Faeces were assayed for the inflammatory marker, lipocalin-2, bacterial loads, and microbiota composition by pyrosequencing. Faecal LPS and flagellin bioactivity were measured via a cell-based reporter assay.
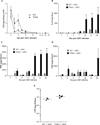
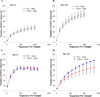
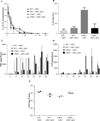
Results: Transient AIEC colonisation, in WT mice, did not alter inflammatory markers, bacterial loads, microbiota composition, nor its pro-inflammatory potential. By contrast, transient AIEC colonisation of T5KO mice drove chronic inflammation which correlated with microbiota components having higher levels of bioactive LPS and flagellin. Such AIEC-induced elevation of LPS and flagellin persisted well beyond AIEC clearance, required AIEC be flagellated, and was associated with alteration in microbiota species composition including a loss of species diversity.
Conclusions: AIEC, and perhaps other pathobionts, may instigate chronic inflammation in susceptible hosts by altering the gut microbiota composition so as to give it an inherently greater ability to activate innate immunity/pro-inflammatory gene expression.
Keywords: COLONIC MICROFLORA; CROHN'S DISEASE; INFLAMMATORY BOWEL DISEASE.
Published by the BMJ Publishing Group Limited. For permission to use (where not already granted under a licence) please go to http://group.bmj.com/group/rights-licensing/permissions.
Figures




Comment in
-
An intestinal arsonist: pathobiont ignites IBD and flees the scene.Gut. 2014 Jul;63(7):1034-5. doi: 10.1136/gutjnl-2013-305589. Epub 2013 Sep 11. Gut. 2014. PMID: 24026350 No abstract available.
References
-
- Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. - PubMed
-
- Chassaing B, Darfeuille-Michaud A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1720–1728. - PubMed
-
- DuPont AW, DuPont HL. The intestinal microbiota and chronic disorders of the gut. Nat Rev Gasto Hepathol. 2011;8:523–531. - PubMed
-
- Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004;127:412–421. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
