BRG1 and BRM chromatin-remodeling complexes regulate the hypoxia response by acting as coactivators for a subset of hypoxia-inducible transcription factor target genes
- PMID: 23897427
- PMCID: PMC3811865
- DOI: 10.1128/MCB.00731-13
BRG1 and BRM chromatin-remodeling complexes regulate the hypoxia response by acting as coactivators for a subset of hypoxia-inducible transcription factor target genes
Abstract
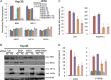
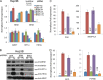
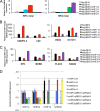
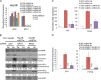
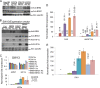
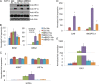
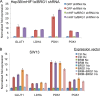
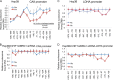
Chromatin remodeling is an active process, which represses or enables the access of transcription machinery to genes in response to external stimuli, including hypoxia. However, in hypoxia, the specific requirement, as well as the molecular mechanism by which the chromatin-remodeling complexes regulate gene expression, remains unclear. In this study, we report that the Brahma (BRM) and Brahma-related gene 1 (BRG1) ATPase-containing SWI/SNF chromatin-remodeling complexes promote the expression of the hypoxia-inducible transcription factor 1α (HIF1α) and HIF2α genes and also promote hypoxic induction of a subset of HIF1 and HIF2 target genes. We show that BRG1 or BRM knockdown in Hep3B and RCC4T cells reduces hypoxic induction of HIF target genes, while reexpression of BRG1 or BRM in BRG1/BRM-deficient SW13 cells increases HIF target gene activation. Mechanistically, HIF1 and HIF2 increase the hypoxic induction of HIF target genes by recruiting BRG1 complexes to HIF target gene promoters, which promotes nucleosome remodeling of HIF target gene promoters in a BRG1 ATPase-dependent manner. Importantly, we found that the function of BRG1 complexes in hypoxic SW13 and RCC4T cells is dictated by the HIF-mediated hypoxia response and could be opposite from their function in normoxic SW13 and RCC4T cells.
Figures


Similar articles
-
Roles of Brahma and Brahma/SWI2-related gene 1 in hypoxic induction of the erythropoietin gene.J Biol Chem. 2004 Nov 5;279(45):46733-41. doi: 10.1074/jbc.M409002200. Epub 2004 Sep 3. J Biol Chem. 2004. PMID: 15347669
-
Heterogeneous SWI/SNF chromatin remodeling complexes promote expression of microphthalmia-associated transcription factor target genes in melanoma.Oncogene. 2010 Jan 7;29(1):81-92. doi: 10.1038/onc.2009.304. Epub 2009 Sep 28. Oncogene. 2010. PMID: 19784067 Free PMC article.
-
Increased activation of the hypoxia-inducible factor pathway in varicose veins.J Vasc Surg. 2012 May;55(5):1427-39. doi: 10.1016/j.jvs.2011.10.111. Epub 2012 Jan 24. J Vasc Surg. 2012. PMID: 22277691
-
The BRG1 ATPase of human SWI/SNF chromatin remodeling enzymes as a driver of cancer.Epigenomics. 2017 Jun;9(6):919-931. doi: 10.2217/epi-2017-0034. Epub 2017 May 19. Epigenomics. 2017. PMID: 28521512 Free PMC article. Review.
-
Involvement of the chromatin-remodeling factor BRG1/SMARCA4 in human cancer.Epigenetics. 2008 Mar-Apr;3(2):64-8. doi: 10.4161/epi.3.2.6153. Epub 2008 Apr 17. Epigenetics. 2008. PMID: 18437052 Review.
Cited by
-
PBRM1 Cooperates with YTHDF2 to Control HIF-1α Protein Translation.Cells. 2021 Jun 8;10(6):1425. doi: 10.3390/cells10061425. Cells. 2021. PMID: 34200988 Free PMC article.
-
The roles of inducible chromatin and transcriptional memory in cellular defense system responses to redox-active pollutants.Free Radic Biol Med. 2021 Jul;170:85-108. doi: 10.1016/j.freeradbiomed.2021.03.018. Epub 2021 Mar 28. Free Radic Biol Med. 2021. PMID: 33789123 Free PMC article. Review.
-
PTPRB promotes metastasis of colorectal carcinoma via inducing epithelial-mesenchymal transition.Cell Death Dis. 2019 Apr 30;10(5):352. doi: 10.1038/s41419-019-1554-9. Cell Death Dis. 2019. PMID: 31040266 Free PMC article.
-
Hypoxia promotes 786-O cells invasiveness and resistance to sorafenib via HIF-2α/COX-2.Med Oncol. 2015 Jan;32(1):419. doi: 10.1007/s12032-014-0419-4. Epub 2014 Dec 7. Med Oncol. 2015. PMID: 25487445
-
Epigenetic regulation of lung cancer cell proliferation and migration by the chromatin remodeling protein BRG1.Oncogenesis. 2019 Nov 6;8(11):66. doi: 10.1038/s41389-019-0174-7. Oncogenesis. 2019. PMID: 31695026 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
