Adaptive strategies and pathogenesis of Clostridium difficile from in vivo transcriptomics
- PMID: 23897605
- PMCID: PMC3811758
- DOI: 10.1128/IAI.00515-13
Adaptive strategies and pathogenesis of Clostridium difficile from in vivo transcriptomics
Erratum in
- Infect Immun. 2014 Feb;82(2):914
Abstract
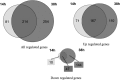
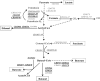
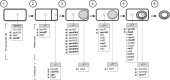
Clostridium difficile is currently the major cause of nosocomial intestinal diseases associated with antibiotic therapy in adults. In order to improve our knowledge of C. difficile-host interactions, we analyzed the genome-wide temporal expression of C. difficile 630 genes during the first 38 h of mouse colonization to identify genes whose expression is modulated in vivo, suggesting that they may play a role in facilitating the colonization process. In the ceca of the C. difficile-monoassociated mice, 549 genes of the C. difficile genome were differentially expressed compared to their expression during in vitro growth, and they were distributed in several functional categories. Overall, our results emphasize the roles of genes involved in host adaptation. Colonization results in a metabolic shift, with genes responsible for the fermentation as well as several other metabolic pathways being regulated inversely to those involved in carbon metabolism. In addition, several genes involved in stress responses, such as ferrous iron uptake or the response to oxidative stress, were regulated in vivo. Interestingly, many genes encoding conserved hypothetical proteins (CHP) were highly and specifically upregulated in vivo. Moreover, genes for all stages of sporulation were quickly induced in vivo, highlighting the observation that sporulation is central to the persistence of C. difficile in the gut and to its ability to spread in the environment. Finally, we inactivated two genes that were differentially expressed in vivo and evaluated the relative colonization fitness of the wild-type and mutant strains in coinfection experiments. We identified a CHP as a putative colonization factor, supporting the suggestion that the in vivo transcriptomic approach can unravel new C. difficile virulence genes.
Figures


Similar articles
-
The flagellin FliC of Clostridium difficile is responsible for pleiotropic gene regulation during in vivo infection.PLoS One. 2014 May 19;9(5):e96876. doi: 10.1371/journal.pone.0096876. eCollection 2014. PLoS One. 2014. PMID: 24841151 Free PMC article.
-
Control of Clostridioides difficile virulence and physiology by the flagellin homeostasis checkpoint FliC-FliW-CsrA in the absence of motility.mBio. 2025 Mar 12;16(3):e0380124. doi: 10.1128/mbio.03801-24. Epub 2025 Jan 30. mBio. 2025. PMID: 39882902 Free PMC article.
-
The Clostridium difficile spo0A gene is a persistence and transmission factor.Infect Immun. 2012 Aug;80(8):2704-11. doi: 10.1128/IAI.00147-12. Epub 2012 May 21. Infect Immun. 2012. PMID: 22615253 Free PMC article.
-
Phase variation of Clostridium difficile virulence factors.Gut Microbes. 2018 Jan 2;9(1):76-83. doi: 10.1080/19490976.2017.1362526. Epub 2017 Sep 21. Gut Microbes. 2018. PMID: 28806147 Free PMC article. Review.
-
Hype or hypervirulence: a reflection on problematic C. difficile strains.Virulence. 2013 Oct 1;4(7):592-6. doi: 10.4161/viru.26297. Epub 2013 Sep 10. Virulence. 2013. PMID: 24060961 Free PMC article. Review.
Cited by
-
Regulating the Intersection of Metabolism and Pathogenesis in Gram-positive Bacteria.Microbiol Spectr. 2015 Jun;3(3):10.1128/microbiolspec.MBP-0004-2014. doi: 10.1128/microbiolspec.MBP-0004-2014. Microbiol Spectr. 2015. PMID: 26185086 Free PMC article. Review.
-
Positive regulation of botulinum neurotoxin gene expression by CodY in Clostridium botulinum ATCC 3502.Appl Environ Microbiol. 2014 Dec;80(24):7651-8. doi: 10.1128/AEM.02838-14. Epub 2014 Oct 3. Appl Environ Microbiol. 2014. PMID: 25281376 Free PMC article.
-
What's a Biofilm?-How the Choice of the Biofilm Model Impacts the Protein Inventory of Clostridioides difficile.Front Microbiol. 2021 Jun 10;12:682111. doi: 10.3389/fmicb.2021.682111. eCollection 2021. Front Microbiol. 2021. PMID: 34177868 Free PMC article.
-
Regulation of Clostridium difficile Spore Formation by the SpoIIQ and SpoIIIA Proteins.PLoS Genet. 2015 Oct 14;11(10):e1005562. doi: 10.1371/journal.pgen.1005562. eCollection 2015 Oct. PLoS Genet. 2015. PMID: 26465937 Free PMC article.
-
Control of Clostridium difficile Infection by Defined Microbial Communities.Microbiol Spectr. 2017 Sep;5(5):10.1128/microbiolspec.bad-0009-2016. doi: 10.1128/microbiolspec.BAD-0009-2016. Microbiol Spectr. 2017. PMID: 28936948 Free PMC article. Review.
References
-
- Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7:526–536 - PubMed
-
- Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, Gerding DN, Gibert M, Popoff MR, Parkhill J, Dougan G, Wren BW. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 10:R102. - PMC - PubMed
-
- Denève C, Janoir C, Poilane I, Fantinato C, Collignon A. 2009. New trends in Clostridium difficile virulence and pathogenesis. Int. J. Antimicrob. Agents 33:S24–S28 - PubMed
-
- La MV, Raoult D, Renesto P. 2008. Regulation of whole bacterial pathogen transcription within infected host. FEMS Microbiol. Rev. 32:440–460 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

