Lgr4 is a key regulator of prostate development and prostate stem cell differentiation
- PMID: 23897697
- PMCID: PMC3934101
- DOI: 10.1002/stem.1484
Lgr4 is a key regulator of prostate development and prostate stem cell differentiation
Abstract
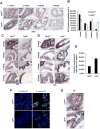
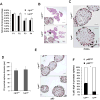
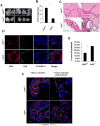
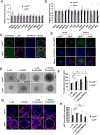
Mechanisms modulating prostate cell fate determination remain unexplored. The leucine-rich repeat containing G-protein-coupled receptors (Lgr) have been identified as important stem cell markers in various tissues. Here, we investigated the roles of Lgr4/Gpr48 in prostate stem cells (PSCs) and development. Lgr4 was ubiquitously expressed during early prostate development prior to lineage specification, with adult expression restricted to a few basal cells (principally Lin(-)Sca1(+)CD49f(+)). Lgr4(-/-) mice had compromised branching morphogenesis and delayed epithelial differentiation, leading to decreased prostate size and impaired luminal cell function. In vitro prostate sphere culture revealed that Lgr4(-/-) Lin(-)/Sca1(+)/CD49f(+) cells failed to generate p63(low) cells, indicating a differentiation deficiency. Furthermore, Lgr4 ablation arrested PSC differentiation of in vivo kidney capsule prostate grafts, suggesting that Lgr4 modulates PSC properties independent of hormonal and mesenchymal effects. Analysis of neonatal prostates and prostate spheres revealed a decrease in Wnt, Sonic Hedgehog, and Notch1 expression in Lgr4(-/-) cells. Lgr4 loss blocked differentiation of prostate sphere p63(hi) cells to p63(low). Treatment with exogenous Sonic Hedgehog partially restored the differentiation of p63(hi) cells in Lgr4(-/-) spheres. Taken together, our data revealed the roles of Lgr4 in early prostate development and in stem cell differentiation through regulation of the Wnt, Notch, and Sonic Hedgehog signaling pathways.
Keywords: Gpr48; Lgr4; Prostate development; Prostate stem cells.
© AlphaMed Press.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Matusik RJ, Jin RJ, Sun Q, et al. Prostate epithelial cell fate. Differentiation; research in biological diversity. 2008;76:682–698. - PubMed
-
- Marker PC. Does prostate cancer co-opt the developmental program? Differentiation; research in biological diversity. 2008;76:736–744. - PubMed
-
- Ousset M, Van Keymeulen A, Bouvencourt G, et al. Multipotent and unipotent progenitors contribute to prostate postnatal development. Nature cell biology. 2012;14:1131–1138. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

