The synaptobrevin homologue Snc2p recruits the exocyst to secretory vesicles by binding to Sec6p
- PMID: 23897890
- PMCID: PMC3734085
- DOI: 10.1083/jcb.201211148
The synaptobrevin homologue Snc2p recruits the exocyst to secretory vesicles by binding to Sec6p
Abstract
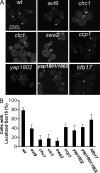
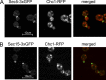
A screen for mutations that affect the recruitment of the exocyst to secretory vesicles identified genes encoding clathrin and proteins that associate or colocalize with clathrin at sites of endocytosis. However, no significant colocalization of the exocyst with clathrin was seen, arguing against a direct role in exocyst recruitment. Rather, these components are needed to recycle the exocytic vesicle SNAREs Snc1p and Snc2p from the plasma membrane into new secretory vesicles where they act to recruit the exocyst. We observe a direct interaction between the exocyst subunit Sec6p and the latter half of the SNARE motif of Snc2p. An snc2 mutation that specifically disrupts this interaction led to exocyst mislocalization and a block in exocytosis in vivo without affecting liposome fusion in vitro. Overexpression of Sec4p partially suppressed the exocyst localization defects of mutations in clathrin and clathrin-associated components. We propose that the exocyst is recruited to secretory vesicles by the combinatorial signals of Sec4-GTP and the Snc proteins. This could help to confer both specificity and directionality to vesicular traffic.
Figures








Similar articles
-
Exocyst stimulates multiple steps of exocytic SNARE complex assembly and vesicle fusion.Nat Struct Mol Biol. 2025 Jan;32(1):150-160. doi: 10.1038/s41594-024-01388-2. Epub 2024 Sep 6. Nat Struct Mol Biol. 2025. PMID: 39242980
-
Functional specialization within a vesicle tethering complex: bypass of a subset of exocyst deletion mutants by Sec1p or Sec4p.J Cell Biol. 2004 Dec 6;167(5):875-87. doi: 10.1083/jcb.200408001. J Cell Biol. 2004. PMID: 15583030 Free PMC article.
-
Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p.J Cell Biol. 2004 Dec 6;167(5):889-901. doi: 10.1083/jcb.200408124. J Cell Biol. 2004. PMID: 15583031 Free PMC article.
-
Dissecting the plant exocyst.Curr Opin Plant Biol. 2019 Dec;52:69-76. doi: 10.1016/j.pbi.2019.08.004. Epub 2019 Sep 8. Curr Opin Plant Biol. 2019. PMID: 31509792 Review.
-
The exocyst complex in polarized exocytosis.Int Rev Cytol. 2004;233:243-65. doi: 10.1016/S0074-7696(04)33006-8. Int Rev Cytol. 2004. PMID: 15037366 Review.
Cited by
-
The role of Sec3p in secretory vesicle targeting and exocyst complex assembly.Mol Biol Cell. 2014 Nov 15;25(23):3813-22. doi: 10.1091/mbc.E14-04-0907. Epub 2014 Sep 17. Mol Biol Cell. 2014. PMID: 25232005 Free PMC article.
-
Polarized Exocytosis.Cold Spring Harb Perspect Biol. 2017 Dec 1;9(12):a027870. doi: 10.1101/cshperspect.a027870. Cold Spring Harb Perspect Biol. 2017. PMID: 28246185 Free PMC article. Review.
-
A quantitative imaging-based screen reveals the exocyst as a network hub connecting endocytosis and exocytosis.Mol Biol Cell. 2015 Jul 1;26(13):2519-34. doi: 10.1091/mbc.E14-11-1527. Epub 2015 May 6. Mol Biol Cell. 2015. PMID: 25947137 Free PMC article.
-
Chaperoning SNARE assembly and disassembly.Nat Rev Mol Cell Biol. 2016 Aug;17(8):465-79. doi: 10.1038/nrm.2016.65. Epub 2016 Jun 15. Nat Rev Mol Cell Biol. 2016. PMID: 27301672 Free PMC article. Review.
-
Exposing the Elusive Exocyst Structure.Trends Biochem Sci. 2018 Sep;43(9):714-725. doi: 10.1016/j.tibs.2018.06.012. Epub 2018 Jul 25. Trends Biochem Sci. 2018. PMID: 30055895 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

