Molecular architecture of plant thylakoids under physiological and light stress conditions: a study of lipid-light-harvesting complex II model membranes
- PMID: 23898030
- PMCID: PMC3723618
- DOI: 10.1105/tpc.113.113076
Molecular architecture of plant thylakoids under physiological and light stress conditions: a study of lipid-light-harvesting complex II model membranes
Abstract
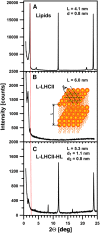
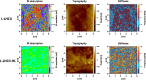
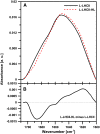
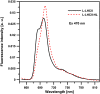
In this study, we analyzed multibilayer lipid-protein membranes composed of the photosynthetic light-harvesting complex II (LHCII; isolated from spinach [Spinacia oleracea]) and the plant lipids monogalcatosyldiacylglycerol and digalactosyldiacylglycerol. Two types of pigment-protein complexes were analyzed: those isolated from dark-adapted leaves (LHCII) and those from leaves preilluminated with high-intensity light (LHCII-HL). The LHCII-HL complexes were found to be partially phosphorylated and contained zeaxanthin. The results of the x-ray diffraction, infrared imaging microscopy, confocal laser scanning microscopy, and transmission electron microscopy revealed that lipid-LHCII membranes assemble into planar multibilayers, in contrast with the lipid-LHCII-HL membranes, which form less ordered structures. In both systems, the protein formed supramolecular structures. In the case of LHCII-HL, these structures spanned the multibilayer membranes and were perpendicular to the membrane plane, whereas in LHCII, the structures were lamellar and within the plane of the membranes. Lamellar aggregates of LHCII-HL have been shown, by fluorescence lifetime imaging microscopy, to be particularly active in excitation energy quenching. Both types of structures were stabilized by intermolecular hydrogen bonds. We conclude that the formation of trans-layer, rivet-like structures of LHCII is an important determinant underlying the spontaneous formation and stabilization of the thylakoid grana structures, since the lamellar aggregates are well suited to dissipate excess energy upon overexcitation.
Figures






References
-
- Ahn T.K., Avenson T.J., Ballottari M., Cheng Y.C., Niyogi K.K., Bassi R., Fleming G.R. (2008). Architecture of a charge-transfer state regulating light harvesting in a plant antenna protein. Science 320: 794–797 - PubMed
-
- Allen J.F. (1992). Protein phosphorylation in regulation of photosynthesis. Biochim. Biophys. Acta 1098: 275–335 - PubMed
-
- Allen J.F. (2003). Botany. State transitions—A question of balance. Science 299: 1530–1532 - PubMed
-
- Barros T., Kuhlbrandt W. (2009). Crystallisation, structure and function of plant light-harvesting complex II. Biochim. Biophys. Acta Bioenerg. 1787: 753–772 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

