ClpS1 is a conserved substrate selector for the chloroplast Clp protease system in Arabidopsis
- PMID: 23898032
- PMCID: PMC3723626
- DOI: 10.1105/tpc.113.112557
ClpS1 is a conserved substrate selector for the chloroplast Clp protease system in Arabidopsis
Abstract
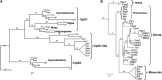
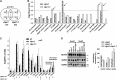
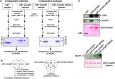
Whereas the plastid caseinolytic peptidase (Clp) P protease system is essential for plant development, substrates and substrate selection mechanisms are unknown. Bacterial ClpS is involved in N-degron substrate selection and delivery to the ClpAP protease. Through phylogenetic analysis, we show that all angiosperms contain ClpS1 and some species also contain ClpS1-like protein(s). In silico analysis suggests that ClpS1 is the functional homolog of bacterial ClpS. We show that Arabidopsis thaliana ClpS1 interacts with plastid ClpC1,2 chaperones. The Arabidopsis ClpS1 null mutant (clps1) lacks a visible phenotype, and no genetic interactions with ClpC/D chaperone or ClpPR core mutants were observed. However, clps1, but not clpc1-1, has increased sensitivity to the translational elongation inhibitor chloramphenicol suggesting a link between translational capacity and ClpS1. Moreover, ClpS1 was upregulated in clpc1-1, and quantitative proteomics of clps1, clpc1, and clps1 clpc1 showed specific molecular phenotypes attributed to loss of ClpC1 or ClpS1. In particular, clps1 showed alteration of the tetrapyrrole pathway. Affinity purification identified eight candidate ClpS1 substrates, including plastid DNA repair proteins and Glu tRNA reductase, which is a control point for tetrapyrrole synthesis. ClpS1 interaction with five substrates strictly depended on two conserved ClpS1 residues involved in N-degron recognition. ClpS1 function, substrates, and substrate recognition mechanisms are discussed.
Figures









References
-
- Bischof S., Baerenfaller K., Wildhaber T., Troesch R., Vidi P.A., Roschitzki B., Hirsch-Hoffmann M., Hennig L., Kessler F., Gruissem W., Baginsky S. (2011). Plastid proteome assembly without Toc159: Photosynthetic protein import and accumulation of N-acetylated plastid precursor proteins. Plant Cell 23: 3911–3928 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

