Random network peristalsis in Physarum polycephalum organizes fluid flows across an individual
- PMID: 23898203
- PMCID: PMC3746869
- DOI: 10.1073/pnas.1305049110
Random network peristalsis in Physarum polycephalum organizes fluid flows across an individual
Abstract
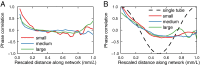
Individuals can function as integrated organisms only when information and resources are shared across a body. Signals and substrates are commonly moved using fluids, often channeled through a network of tubes. Peristalsis is one mechanism for fluid transport and is caused by a wave of cross-sectional contractions along a tube. We extend the concept of peristalsis from the canonical case of one tube to a random network. Transport is maximized within the network when the wavelength of the peristaltic wave is of the order of the size of the network. The slime mold Physarum polycephalum grows as a random network of tubes, and our experiments confirm peristalsis is used by the slime mold to drive internal cytoplasmic flows. Comparisons of theoretically generated contraction patterns with the patterns exhibited by individuals of P. polycephalum demonstrate that individuals maximize internal flows by adapting patterns of contraction to size, thus optimizing transport throughout an organism. This control of fluid flow may be the key to coordinating growth and behavior, including the dynamic changes in network architecture seen over time in an individual.
Keywords: acellular; fungi; myxomycete.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Boddy L, Hynes J, Bebber DP, Fricker MD. Saprotrophic cord systems: Dispersal mechanisms in space and time. Mycoscience. 2009;50(1):9–19.
-
- Nakagaki T, Guy R. Intelligent behaviors of amoeboid movement based on complex dynamics of soft matter. Soft Matter. 2008;4(1):57–67. - PubMed
-
- Nakagaki T, et al. Minimum-risk path finding by an adaptive amoebal network. Phys Rev Lett. 2007;99(6):068104. - PubMed
-
- Nakagaki T, Yamada H, Tóth A. Maze-solving by an amoeboid organism. Nature. 2000;407(6803):470. - PubMed
-
- Tero A, et al. Rules for biologically inspired adaptive network design. Science. 2010;327(5964):439–442. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

