The iron-sulfur cluster assembly machineries in plants: current knowledge and open questions
- PMID: 23898337
- PMCID: PMC3721309
- DOI: 10.3389/fpls.2013.00259
The iron-sulfur cluster assembly machineries in plants: current knowledge and open questions
Abstract
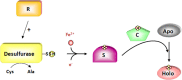
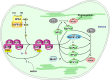
Many metabolic pathways and cellular processes occurring in most sub-cellular compartments depend on the functioning of iron-sulfur (Fe-S) proteins, whose cofactors are assembled through dedicated protein machineries. Recent advances have been made in the knowledge of the functions of individual components through a combination of genetic, biochemical and structural approaches, primarily in prokaryotes and non-plant eukaryotes. Whereas most of the components of these machineries are conserved between kingdoms, their complexity is likely increased in plants owing to the presence of additional assembly proteins and to the existence of expanded families for several assembly proteins. This review focuses on the new actors discovered in the past few years, such as glutaredoxin, BOLA and NEET proteins as well as MIP18, MMS19, TAH18, DRE2 for the cytosolic machinery, which are integrated into a model for the plant Fe-S cluster biogenesis systems. It also discusses a few issues currently subjected to an intense debate such as the role of the mitochondrial frataxin and of glutaredoxins, the functional separation between scaffold, carrier and iron-delivery proteins and the crosstalk existing between different organelles.
Keywords: assembly machineries; carrier Proteins; iron donor; iron-sulfur; repair; scaffold proteins.
Figures




References
-
- Angelini S., Gerez C., Ollagnier-de Choudens S., Sanakis Y., Fontecave M., Barras F., et al. (2008). NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J. Biol. Chem. 283, 14084–14091 10.1074/jbc.M709405200 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

