A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics
- PMID: 23898399
- PMCID: PMC3721235
- DOI: 10.7554/eLife.00762
A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics
Abstract
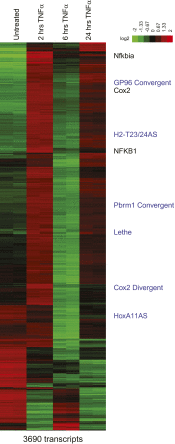
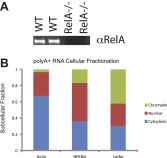
Pseudogenes are thought to be inactive gene sequences, but recent evidence of extensive pseudogene transcription raised the question of potential function. Here we discover and characterize the sets of mouse lncRNAs induced by inflammatory signaling via TNFα. TNFα regulates hundreds of lncRNAs, including 54 pseudogene lncRNAs, several of which show exquisitely selective expression in response to specific cytokines and microbial components in a NF-κB-dependent manner. Lethe, a pseudogene lncRNA, is selectively induced by proinflammatory cytokines via NF-κB or glucocorticoid receptor agonist, and functions in negative feedback signaling to NF-κB. Lethe interacts with NF-κB subunit RelA to inhibit RelA DNA binding and target gene activation. Lethe level decreases with organismal age, a physiological state associated with increased NF-κB activity. These findings suggest that expression of pseudogenes lncRNAs are actively regulated and constitute functional regulators of inflammatory signaling. DOI:http://dx.doi.org/10.7554/eLife.00762.001.
Keywords: Mouse; NF-κB; genomics; noncoding RNA.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

