Multiplexed electrochemistry of DNA-bound metalloproteins
- PMID: 23899026
- PMCID: PMC3899834
- DOI: 10.1021/ja4041779
Multiplexed electrochemistry of DNA-bound metalloproteins
Abstract
Here we describe a multiplexed electrochemical characterization of DNA-bound proteins containing [4Fe-4S] clusters. DNA-modified electrodes have become an essential tool for the characterization of the redox chemistry of DNA repair proteins containing redox cofactors, and multiplexing offers a means to probe different complex samples and substrates in parallel to elucidate this chemistry. Multiplexed analysis of endonuclease III (EndoIII), a DNA repair protein containing a [4Fe-4S] cluster known to be accessible via DNA-mediated charge transport, shows subtle differences in the electrochemical behavior as a function of DNA morphology. The peak splitting, signal broadness, sensitivity to π-stack perturbations, and kinetics were all characterized for the DNA-bound reduction of EndoIII on both closely and loosely packed DNA films. DNA-bound EndoIII is seen to have two different electron transfer pathways for reduction, either through the DNA base stack or through direct surface reduction; closely packed DNA films, where the protein has limited surface accessibility, produce electrochemical signals reflecting electron transfer that is DNA-mediated. Multiplexing furthermore permits the comparison of the electrochemistry of EndoIII mutants, including a new family of mutations altering the electrostatics surrounding the [4Fe-4S] cluster. While little change in the midpoint potential was found for this family of mutants, significant variations in the efficiency of DNA-mediated electron transfer were apparent. On the basis of the stability of these proteins, examined by circular dichroism, we propose that the electron transfer pathway can be perturbed not only by the removal of aromatic residues but also through changes in solvation near the cluster.
Figures



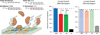



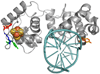
References
-
- Zheng G, Patolsky F, Cui Y, Wang UW, Lieber CM. Nature Biotechnol. 2005;23:1294. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

