An ER-peroxisome tether exerts peroxisome population control in yeast
- PMID: 23900285
- PMCID: PMC3770948
- DOI: 10.1038/emboj.2013.170
An ER-peroxisome tether exerts peroxisome population control in yeast
Abstract
Eukaryotic cells compartmentalize biochemical reactions into membrane-enclosed organelles that must be faithfully propagated from one cell generation to the next. Transport and retention processes balance the partitioning of organelles between mother and daughter cells. Here we report the identification of an ER-peroxisome tether that links peroxisomes to the ER and ensures peroxisome population control in the yeast Saccharomyces cerevisiae. The tether consists of the peroxisome biogenic protein, Pex3p, and the peroxisome inheritance factor, Inp1p. Inp1p bridges the two compartments by acting as a molecular hinge between ER-bound Pex3p and peroxisomal Pex3p. Asymmetric peroxisome division leads to the formation of Inp1p-containing anchored peroxisomes and Inp1p-deficient mobile peroxisomes that segregate to the bud. While peroxisomes in mother cells are not released from tethering, de novo formation of tethers in the bud assists in the directionality of peroxisome transfer. Peroxisomes are thus stably maintained over generations of cells through their continued interaction with tethers.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


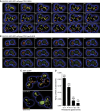

Comment in
-
Give what you can and keep what you need!EMBO J. 2013 Sep 11;32(18):2424-6. doi: 10.1038/emboj.2013.180. Epub 2013 Aug 6. EMBO J. 2013. PMID: 23921554 Free PMC article.
References
-
- Fagarasanu A, Fagarasanu M, Eitzen GA, Aitchison JD, Rachubinski RA (2006) The peroxisomal membrane protein Inp2p of the peroxisome-specific receptor for the myosin V motor Myo2p of Saccharomyces cerevisiae. Dev Cell 10: 587–600 - PubMed
-
- Fagarasanu A, Mast FD, Knoblach B, Rachubinski RA (2010) Molecular mechanisms of organelle inheritance: lessons from peroxisomes in yeast. Nat Rev Mol Cell Biol 11: 644–654 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

