Mobilization of a plant transposon by expression of the transposon-encoded anti-silencing factor
- PMID: 23900287
- PMCID: PMC3773815
- DOI: 10.1038/emboj.2013.169
Mobilization of a plant transposon by expression of the transposon-encoded anti-silencing factor
Abstract
Transposable elements (TEs) have a major impact on genome evolution, but they are potentially deleterious, and most of them are silenced by epigenetic mechanisms, such as DNA methylation. Here, we report the characterization of a TE encoding an activity to counteract epigenetic silencing by the host. In Arabidopsis thaliana, we identified a mobile copy of the Mutator-like element (MULE) with degenerated terminal inverted repeats (TIRs). This TE, named Hiun (Hi), is silent in wild-type plants, but it transposes when DNA methylation is abolished. When a Hi transgene was introduced into the wild-type background, it induced excision of the endogenous Hi copy, suggesting that Hi is the autonomously mobile copy. In addition, the transgene induced loss of DNA methylation and transcriptional activation of the endogenous Hi. Most importantly, the trans-activation of Hi depends on a Hi-encoded protein different from the conserved transposase. Proteins related to this anti-silencing factor, which we named VANC, are widespread in the non-TIR MULEs and may have contributed to the recent success of these TEs in natural Arabidopsis populations.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

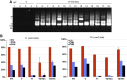







 , where Mn, Cn, Mt and Ct are methylated cytosine (M) and total cytosine (C) counts mapped for each TE in the non-transgenic (n) and transgenic (t) plants, respectively. Each of these values is shown in Supplementary Table S3, with the corresponding values for CpG sites. This figure contains 24 282 TEs plotted, while 6910 TEs are not plotted due to lack of mapped cytosine in one or more of the three contexts in non-transgenic or transgenic plant. The lack is mainly because we did not use reads mapped multiple loci. Some of the VANDAL21 family members (surrounded by broken ellipse) showed most significant reduction for both CpHpG and CpHpH sites. The dot for Hi is indicated by an arrow. Similar analyses done on genes are shown in Supplementary Figure S12. (B) Effect of Hi transgene on DNA methylation across each of the VANDAL21 members. Left and right terminals are shown by broken lines for each element. Each point represents proportion of methylated cytosine for a sliding window with seven fractions after separating each TE for 100 fractions. Right and left flanking regions are also analysed by the same conditions. Scale bars for CpG, CpHpG, and CpHpH sites in each panel indicate 1, 0.8, and 0.4, respectively. For some of the VANDAL21 copies, reduced methylation in the terminal region is also confirmed by conventional bisulphite sequencing using primers within and flanking the TE (Supplementary Figure S13). Results of DNA methylation across six additional VANDAL21 members are shown in Supplementary Figure S14. CACTA2 is shown as a negative control.
, where Mn, Cn, Mt and Ct are methylated cytosine (M) and total cytosine (C) counts mapped for each TE in the non-transgenic (n) and transgenic (t) plants, respectively. Each of these values is shown in Supplementary Table S3, with the corresponding values for CpG sites. This figure contains 24 282 TEs plotted, while 6910 TEs are not plotted due to lack of mapped cytosine in one or more of the three contexts in non-transgenic or transgenic plant. The lack is mainly because we did not use reads mapped multiple loci. Some of the VANDAL21 family members (surrounded by broken ellipse) showed most significant reduction for both CpHpG and CpHpH sites. The dot for Hi is indicated by an arrow. Similar analyses done on genes are shown in Supplementary Figure S12. (B) Effect of Hi transgene on DNA methylation across each of the VANDAL21 members. Left and right terminals are shown by broken lines for each element. Each point represents proportion of methylated cytosine for a sliding window with seven fractions after separating each TE for 100 fractions. Right and left flanking regions are also analysed by the same conditions. Scale bars for CpG, CpHpG, and CpHpH sites in each panel indicate 1, 0.8, and 0.4, respectively. For some of the VANDAL21 copies, reduced methylation in the terminal region is also confirmed by conventional bisulphite sequencing using primers within and flanking the TE (Supplementary Figure S13). Results of DNA methylation across six additional VANDAL21 members are shown in Supplementary Figure S14. CACTA2 is shown as a negative control.References
-
- Brettell RI, Dennis ES (1991) Reactivation of a silent Ac following tissue culture is associated with heritable alterations in its methylation. Mol Gen Genet 229: 365–372 - PubMed
-
- Buisine N, Quesneville H, Colot V (2008) Improved detection and annotation of transposable elements in sequenced genomes using multiple reference sequence sets. Genomics 91: 467–475 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

