Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity
- PMID: 23901100
- PMCID: PMC3746923
- DOI: 10.1073/pnas.1303781110
Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity
Abstract
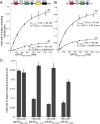
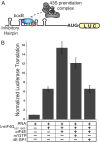
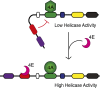
Elevated eukaryotic initiation factor 4E (eIF4E) levels frequently occur in a variety of human cancers. Overexpression of eIF4E promotes cellular transformation by selectively increasing the translation of proliferative and prosurvival mRNAs. These mRNAs possess highly structured 5'-UTRs that impede ribosome recruitment and scanning, yet the mechanism for how eIF4E abundance elevates their translation is not easily explained by its cap-binding activity. Here, we show that eIF4E possesses an unexpected second function in translation initiation by strongly stimulating eukaryotic initiation factor 4A (eIF4A) helicase activity. Importantly, we demonstrate that this activity promotes mRNA restructuring in a manner that is independent of its cap-binding function. To explain these findings, we show that the eIF4E-binding site in eukaryotic initiation factor 4G (eIF4G) functions as an autoinhibitory domain to modulate its ability to stimulate eIF4A helicase activity. Binding of eIF4E counteracts this autoinhibition, enabling eIF4G to stimulate eIF4A helicase activity. Finally, we have successfully separated the two functions of eIF4E to show that its helicase promoting activity increases the rate of translation by a mechanism that is distinct from its cap-binding function. Based on our results, we propose that maintaining a connection between eIF4E and eIF4G throughout scanning provides a plausible mechanism to explain how eIF4E abundance selectively stimulates the translation of highly structured proliferation and tumor-promoting mRNAs.
Keywords: ATPase; DEAD-box; protein synthesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10(4):254–266. - PubMed
-
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. - PubMed
-
- Duncan R, Milburn SC, Hershey JW. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J Biol Chem. 1987;262(1):380–388. - PubMed
-
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433(7025):477–480. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

