Combinatorial regulation of meiotic holliday junction resolution in C. elegans by HIM-6 (BLM) helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 nucleases
- PMID: 23901331
- PMCID: PMC3715425
- DOI: 10.1371/journal.pgen.1003591
Combinatorial regulation of meiotic holliday junction resolution in C. elegans by HIM-6 (BLM) helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 nucleases
Erratum in
- PLoS Genet. 2013 Aug;9(8). doi:10.1371/annotation/d8c73205-151d-4e22-89c6-3aa574037d10
Abstract
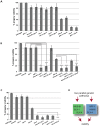
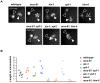
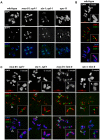
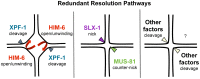
Holliday junctions (HJs) are cruciform DNA structures that are created during recombination events. It is a matter of considerable importance to determine the resolvase(s) that promote resolution of these structures. We previously reported that C. elegans GEN-1 is a symmetrically cleaving HJ resolving enzyme required for recombinational repair, but we could not find an overt role in meiotic recombination. Here we identify C. elegans proteins involved in resolving meiotic HJs. We found no evidence for a redundant meiotic function of GEN-1. In contrast, we discovered two redundant HJ resolution pathways likely coordinated by the SLX-4 scaffold protein and also involving the HIM-6/BLM helicase. SLX-4 associates with the SLX-1, MUS-81 and XPF-1 nucleases and has been implicated in meiotic recombination in C. elegans. We found that C. elegans [mus-81; xpf-1], [slx-1; xpf-1], [mus-81; him-6] and [slx-1; him-6] double mutants showed a similar reduction in survival rates as slx-4. Analysis of meiotic diakinesis chromosomes revealed a distinct phenotype in these double mutants. Instead of wild-type bivalent chromosomes, pairs of "univalents" linked by chromatin bridges occur. These linkages depend on the conserved meiosis-specific transesterase SPO-11 and can be restored by ionizing radiation, suggesting that they represent unresolved meiotic HJs. This suggests the existence of two major resolvase activities, one provided by XPF-1 and HIM-6, the other by SLX-1 and MUS-81. In all double mutants crossover (CO) recombination is reduced but not abolished, indicative of further redundancy in meiotic HJ resolution. Real time imaging revealed extensive chromatin bridges during the first meiotic division that appear to be eventually resolved in meiosis II, suggesting back-up resolution activities acting at or after anaphase I. We also show that in HJ resolution mutants, the restructuring of chromosome arms distal and proximal to the CO still occurs, suggesting that CO initiation but not resolution is likely to be required for this process.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Comment in
-
An elegans Solution for Crossover Formation.PLoS Genet. 2013;9(7):e1003658. doi: 10.1371/journal.pgen.1003658. Epub 2013 Jul 18. PLoS Genet. 2013. PMID: 23874241 Free PMC article. No abstract available.
References
-
- Sun H, Treco D, Schultes NP, Szostak JW (1989) Double-strand breaks at an initiation site for meiotic gene conversion. Nature 338: 87–90. - PubMed
-
- Keeney S, Giroux CN, Kleckner N (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. - PubMed
-
- Moens PB, Kolas NK, Tarsounas M, Marcon E, Cohen PE, et al. (2002) The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J Cell Sci 115: 1611–1622. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

