Pyrimidine homeostasis is accomplished by directed overflow metabolism
- PMID: 23903661
- PMCID: PMC4470420
- DOI: 10.1038/nature12445
Pyrimidine homeostasis is accomplished by directed overflow metabolism
Abstract
Cellular metabolism converts available nutrients into usable energy and biomass precursors. The process is regulated to facilitate efficient nutrient use and metabolic homeostasis. Feedback inhibition of the first committed step of a pathway by its final product is a classical means of controlling biosynthesis. In a canonical example, the first committed enzyme in the pyrimidine pathway in Escherichia coli is allosterically inhibited by cytidine triphosphate. The physiological consequences of disrupting this regulation, however, have not been previously explored. Here we identify an alternative regulatory strategy that enables precise control of pyrimidine pathway end-product levels, even in the presence of dysregulated biosynthetic flux. The mechanism involves cooperative feedback regulation of the near-terminal pathway enzyme uridine monophosphate kinase. Such feedback leads to build-up of the pathway intermediate uridine monophosphate, which is in turn degraded by a conserved phosphatase, here termed UmpH, with previously unknown physiological function. Such directed overflow metabolism allows homeostasis of uridine triphosphate and cytidine triphosphate levels at the expense of uracil excretion and slower growth during energy limitation. Disruption of the directed overflow regulatory mechanism impairs growth in pyrimidine-rich environments. Thus, pyrimidine homeostasis involves dual regulatory strategies, with classical feedback inhibition enhancing metabolic efficiency and directed overflow metabolism ensuring end-product homeostasis.
Conflict of interest statement
Figures


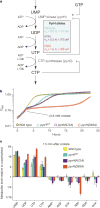
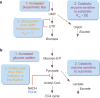
Comment in
-
Biochemistry: Curbing the excesses of low demand.Nature. 2013 Aug 8;500(7461):157-8. doi: 10.1038/nature12461. Epub 2013 Jul 31. Nature. 2013. PMID: 23903659 No abstract available.
References
-
- Gerhart JC, Pardee AB. The enzymology of control by feedback inhibition. J Biol Chem. 1962;237:891–896. - PubMed
-
- Savageau MA. Optimal design of feedback-control by inhibition — dynamic considerations. J Mol Evol. 1975;5:199–222. - PubMed
-
- Umbarger HE. Evidence for a negative-feedback mechanism in the biosynthesis of isoleucine. Science. 1956;123:848–849. - PubMed
-
- Pardee AB, Yates RA. Control of pyrimidine biosynthesis in Escherichia coli by a feed-back mechanism. J Biol Chem. 1956;221:757–770. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

