Genetic burden in multiple sclerosis families
- PMID: 23903824
- PMCID: PMC4102601
- DOI: 10.1038/gene.2013.37
Genetic burden in multiple sclerosis families
Abstract
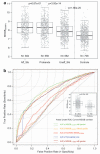
A previous study using cumulative genetic risk estimations in multiple sclerosis (MS) successfully tracked the aggregation of susceptibility variants in multi-case and single-case families. It used a limited description of susceptibility loci available at the time (17 loci). Even though the full roster of MS risk genes remains unavailable, we estimated the genetic burden in MS families and assess its disease predictive power using up to 64 single-nucleotide polymorphism (SNP) markers according to the most recent literature. A total of 708 controls, 3251 MS patients and their relatives, as well as 117 twin pairs were genotyped. We validated the increased aggregation of genetic burden in multi-case compared with single-case families (P=4.14e-03) and confirm that these data offer little opportunity to accurately predict MS, even within sibships (area under receiver operating characteristic (AUROC)=0.59 (0.55, 0.53)). Our results also suggest that the primary progressive and relapsing-type forms of MS share a common genetic architecture (P=0.368; difference being limited to that corresponding to ± 2 typical MS-associated SNPs). We have confirmed the properties of individual genetic risk score in MS. Comparing with previous reference point for MS genetics (17 SNPs), we underlined the corrective consequences of the integration of the new findings from GWAS and meta-analysis.
Figures



References
-
- Oksenberg JR, Baranzini SE, Sawcer S, Hauser SL. The genetics of multiple sclerosis: SNPs to pathways to pathogenesis. Nat Rev Genet. 2008;9:516–526. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

