Isolate-specific differences in the conformational dynamics and antigenicity of HIV-1 gp120
- PMID: 23903848
- PMCID: PMC3807424
- DOI: 10.1128/JVI.01535-13
Isolate-specific differences in the conformational dynamics and antigenicity of HIV-1 gp120
Abstract
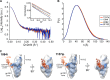
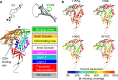
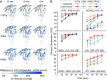
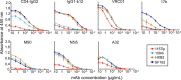
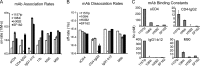
The HIV-1 envelope glycoprotein (Env) mediates viral entry into host cells and is the sole target of neutralizing antibodies. Much of the sequence diversity in the HIV-1 genome is concentrated within Env, particularly within its gp120 surface subunit. While dramatic functional diversity exists among HIV-1 Env isolates-observable even in the context of monomeric gp120 proteins as differences in antigenicity and immunogenicity-we have little understanding of the structural features that distinguish Env isolates and lead to isolate-specific functional differences, as crystal structures of truncated gp120 "core" proteins from diverse isolates reveal a high level of structural conservation. Because gp120 proteins are used as prospective vaccine immunogens, it is critical to understand the structural factors that influence their reactivity with antibodies. Here, we studied four full-length, glycosylated gp120 monomers from diverse HIV-1 isolates by using small-angle X-ray scattering (SAXS) to probe the overall subunit morphology and hydrogen/deuterium-exchange with mass spectrometry (HDX-MS) to characterize the local structural order of each gp120. We observed that while the overall subunit architecture was similar among isolates by SAXS, dramatic isolate-specific differences in the conformational stability of gp120 were evident by HDX-MS. These differences persisted even with the CD4 receptor bound. Furthermore, surface plasmon resonance (SPR) and enzyme-linked immunosorbance assays (ELISAs) showed that disorder was associated with poorer recognition by antibodies targeting conserved conformational epitopes. These data provide additional insight into the structural determinants of gp120 antigenicity and suggest that conformational dynamics should be considered in the selection and design of optimized Env immunogens.
Figures





References
-
- Allan JS, Coligan JE, Barin F, McLane MF, Sodroski JG, Rosen CA, Haseltine WA, Lee TH, Essex M. 1985. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science 228:1091–1094 - PubMed
-
- Lasky LA, Groopman JE, Fennie CW, Benz PM, Capon DJ, Dowbenko DJ, Nakamura GR, Nunes WM, Renz ME, Berman PW. 1986. Neutralization of the AIDS retrovirus by antibodies to a recombinant envelope glycoprotein. Science 233:209–212 - PubMed
-
- Barin F, McLane MF, Allan JS, Lee TH, Groopman JE, Essex M. 1985. Virus envelope protein of HTLV-III represents major target antigen for antibodies in AIDS patients. Science 228:1094–1096 - PubMed
-
- Hoxie JA. 2010. Toward an antibody-based HIV-1 vaccine. Annu. Rev. Med. 61:135–152 - PubMed
-
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01-GM099989/GM/NIGMS NIH HHS/United States
- R00 GM080352/GM/NIGMS NIH HHS/United States
- R01 AI076170/AI/NIAID NIH HHS/United States
- P51 RR000166/RR/NCRR NIH HHS/United States
- R00-GM080352/GM/NIGMS NIH HHS/United States
- P41 RR001209/RR/NCRR NIH HHS/United States
- P41-RR001209/RR/NCRR NIH HHS/United States
- T32-AI7509/AI/NIAID NIH HHS/United States
- P01-RR000166/RR/NCRR NIH HHS/United States
- P30-AI027767/AI/NIAID NIH HHS/United States
- P30 AI027767/AI/NIAID NIH HHS/United States
- T32 AI007509/AI/NIAID NIH HHS/United States
- R01 GM099989/GM/NIGMS NIH HHS/United States
- R01-AI076170/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

