The genomic map of breast cancer: which roads lead to better targeted therapies?
- PMID: 23905624
- PMCID: PMC3979080
- DOI: 10.1186/bcr3435
The genomic map of breast cancer: which roads lead to better targeted therapies?
Abstract
Recent advances in whole-genome technologies have supplied the field of cancer research with an overwhelming amount of molecular data. Improvements in massively parallel sequencing approaches have led to logarithmic decreases in costs, and so these methods are becoming almost commonplace in the analysis of clinical trials and other cohorts of interest. Furthermore, whole-transcriptome quantification by RNA sequencing is quickly replacing microarrays. However, older chip-based methodologies such as comparative genomic hybridization and single-nucleotide polymorphism arrays have benefited from this technological explosion and are now so accessible that they can be employed in increasingly larger cohorts of patients. The study of breast cancer lends itself particularly well to these technologies. It is the most commonly diagnosed neoplasm in women, giving rise to nearly 230,000 new cases each year. Many patients are given a diagnosis of early-stage disease, for which surgery is the standard of care. These attributes result in excellent availability of tissues for whole-genome/transcriptome analysis. The Cancer Genome Atlas project has generated comprehensive catalogs of publically available genomic breast cancer data. In addition, other studies employing the power of genomic technologies in medium to large cohorts were recently published. These data are now publically available for the generation of novel hypotheses. However, these studies differed in the methods, patient cohorts, and analytical techniques employed and represent complementary snapshots of the molecular underpinnings of breast cancer. Here, we will discuss the convergences and divergences of these reports as well as the scientific and clinical implications of their findings.
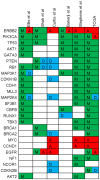
Figures
References
-
- Rakha EA, Reis-Filho JS, Baehner F, Dabbs DJ, Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR, Palacios J, Richardson AL, Schnitt SJ, Schmitt FC, Tan PH, Tse GM, Badve S, Ellis IO. Breast cancer prognostic classification in the molecular era: the role of histological grade. Breast Cancer Res. 2010;15:207. - PMC - PubMed
-
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;15:1160–1167. doi: 10.1200/JCO.2008.18.1370. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

