CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering
- PMID: 23907171
- PMCID: PMC3818127
- DOI: 10.1038/nbt.2675
CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering
Abstract
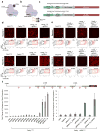
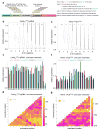
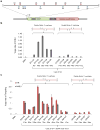
Prokaryotic type II CRISPR-Cas systems can be adapted to enable targeted genome modifications across a range of eukaryotes. Here we engineer this system to enable RNA-guided genome regulation in human cells by tethering transcriptional activation domains either directly to a nuclease-null Cas9 protein or to an aptamer-modified single guide RNA (sgRNA). Using this functionality we developed a transcriptional activation-based assay to determine the landscape of off-target binding of sgRNA:Cas9 complexes and compared it with the off-target activity of transcription activator-like (TALs) effectors. Our results reveal that specificity profiles are sgRNA dependent, and that sgRNA:Cas9 complexes and 18-mer TAL effectors can potentially tolerate 1-3 and 1-2 target mismatches, respectively. By engineering a requirement for cooperativity through offset nicking for genome editing or through multiple synergistic sgRNAs for robust transcriptional activation, we suggest methods to mitigate off-target phenomena. Our results expand the versatility of the sgRNA:Cas9 tool and highlight the critical need to engineer improved specificity.
Figures
Comment in
-
Staying on target with CRISPR-Cas.Nat Biotechnol. 2013 Sep;31(9):807-9. doi: 10.1038/nbt.2684. Nat Biotechnol. 2013. PMID: 24022156 No abstract available.
References
-
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature biotechnology. 2013;31:230–232. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

