Suppression of PTRF alleviates the polymicrobial sepsis induced by cecal ligation and puncture in mice
- PMID: 23908488
- PMCID: PMC3814834
- DOI: 10.1093/infdis/jit364
Suppression of PTRF alleviates the polymicrobial sepsis induced by cecal ligation and puncture in mice
Abstract
Background: Sepsis and sepsis-associated organ failure are devastating conditions. Understanding the detailed cellular/molecular mechanisms involved in sepsis should lead to the identification of novel therapeutic targets.
Methods: Cecal ligation and puncture (CLP) was used as a polymicrobial sepsis model in vivo to determine mortality and end-organ damage. Macrophages were adopted as the cellular model in vitro for mechanistic studies.
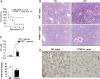
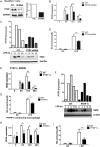
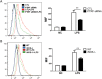
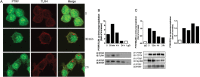
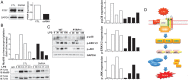
Results: PTRF+/- mice survived longer and suffered less organ damage after CLP. Reductions in nitric oxide (NO) and iNOS biosynthesis were observed in plasma, macrophages, and vital organs in the PTRF+/- mice. Using an acute sepsis model after CLP, we found that iNOS-/- mice had a comparable level of survival as the PTRF+/- mice. Similarly, polymerase I transcript release factor (PTRF) deficiency resulted in decreased iNOS and NO/ROS production in macrophages in vitro. Mechanistically, lipopolysaccharide (LPS) enhanced the co-localization and interaction between PTRF and TLR4 in lipid rafts. Deletion of PTRF blocked formation of the TLR4/Myd88 complex after LPS. Consistent with this, lack of PTRF impaired the TLR4 signaling, as shown by the decreased p-JNK, p-ERK, and p-p38, which are upstream factors involved in iNOS transcription.
Conclusion: PTRF is a crucial regulator of TLR4 signaling in the development of sepsis.
Keywords: CLP; PTRF; ROS; TLR4; macrophage; nitric oxide; sepsis.
Figures


Similar articles
-
Reduning injection and its effective constituent luteoloside protect against sepsis partly via inhibition of HMGB1/TLR4/NF-κB/MAPKs signaling pathways.J Ethnopharmacol. 2021 Apr 24;270:113783. doi: 10.1016/j.jep.2021.113783. Epub 2021 Jan 7. J Ethnopharmacol. 2021. PMID: 33421596
-
Lipopolysaccharide clearance, bacterial clearance, and systemic inflammatory responses are regulated by cell type-specific functions of TLR4 during sepsis.J Immunol. 2013 May 15;190(10):5152-60. doi: 10.4049/jimmunol.1300496. Epub 2013 Apr 5. J Immunol. 2013. PMID: 23562812 Free PMC article.
-
Gene deletion of protein tyrosine phosphatase 1B protects against sepsis-induced cardiovascular dysfunction and mortality.Arterioscler Thromb Vasc Biol. 2014 May;34(5):1032-44. doi: 10.1161/ATVBAHA.114.303450. Epub 2014 Feb 27. Arterioscler Thromb Vasc Biol. 2014. PMID: 24578383
-
Cecal ligation and puncture-induced sepsis as a model to study autophagy in mice.J Vis Exp. 2014 Feb 9;(84):e51066. doi: 10.3791/51066. J Vis Exp. 2014. PMID: 24561344 Free PMC article. Review.
-
Evaluation of the Molecular Mechanisms of Sepsis Using Proteomics.Front Immunol. 2021 Oct 21;12:733537. doi: 10.3389/fimmu.2021.733537. eCollection 2021. Front Immunol. 2021. PMID: 34745104 Free PMC article. Review.
Cited by
-
Molecular and clinical characterization of PTRF in glioma via 1,022 samples.BMC Cancer. 2023 Jun 16;23(1):551. doi: 10.1186/s12885-023-11001-2. BMC Cancer. 2023. PMID: 37322408 Free PMC article.
-
Cavin-1 promotes M2 macrophages/microglia polarization via SOCS3.Inflamm Res. 2022 Apr;71(4):397-407. doi: 10.1007/s00011-022-01550-w. Epub 2022 Mar 11. Inflamm Res. 2022. PMID: 35275225
-
Immune response and differentially expressed proteins in the lung tissue of BALB/c mice challenged by aerosolized Brucella melitensis 5.J Int Med Res. 2018 Nov;46(11):4740-4752. doi: 10.1177/0300060518799879. Epub 2018 Oct 3. J Int Med Res. 2018. PMID: 30282518 Free PMC article.
-
Caveolae and the oxidative stress response.Biochem Soc Trans. 2023 Jun 28;51(3):1377-1385. doi: 10.1042/BST20230121. Biochem Soc Trans. 2023. PMID: 37248872 Free PMC article.
-
Suppression of PTRF Alleviates Post-Infectious Irritable Bowel Syndrome via Downregulation of the TLR4 Pathway in Rats.Front Pharmacol. 2021 Oct 7;12:724410. doi: 10.3389/fphar.2021.724410. eCollection 2021. Front Pharmacol. 2021. PMID: 34690766 Free PMC article.
References
-
- Levy MM, Fink MP, Marshall JC, et al. 2001 Sccm/Esicm/Accp/Ats/Sis international sepsis definitions conference. Crit Care Med. 2003;31:1250–6. - PubMed
-
- Munford RS. Severe sepsis and septic shock: the role of gram-negative bacteremia. Annu Rev Pathol. 2006;1:467–96. - PubMed
-
- Annane D, Bellissant E, Cavaillon JM. Septic shock. Lancet. 2005;365:63–78. - PubMed
-
- Biswas SK, Chittezhath M, Shalova IN, Lim JY. Macrophage polarization and plasticity in health and disease. Immunol Res. 2012;53:11–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

