Mass spectrometry-based analysis of rat liver and hepatocellular carcinoma Morris hepatoma 7777 plasma membrane proteome
- PMID: 23909495
- PMCID: PMC3840720
- DOI: 10.1021/ac400774g
Mass spectrometry-based analysis of rat liver and hepatocellular carcinoma Morris hepatoma 7777 plasma membrane proteome
Abstract
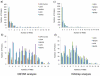
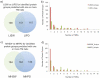
The gel-based proteomic analysis of plasma membranes from rat liver and chemically induced, malignant hepatocellular carcinoma Morris hepatoma 7777 was systematically optimized to yield the maximum number of proteins containing transmembrane domains (TMDs). Incorporation of plasma membrane proteins into a polyacrylamide "tube gel" followed by in-gel digestion of "tube gel" pieces significantly improved detection by electrospray ionization-liquid chromatography-tandem mass spectrometry. Removal of less hydrophobic proteins by washing isolated plasma membranes with 0.1 M sodium carbonate enables detection of a higher number of hydrophobic proteins containing TMDs in both tissues. Subsequent treatment of plasma membranes by a proteolytic enzyme (trypsin) causes the loss of some of the proteins that are detected after washing with sodium carbonate, but it enables the detection of other hydrophobic proteins containing TMDs. Introduction of mass spectrometers with higher sensitivity, higher mass resolution and mass accuracy, and a faster scan rate significantly improved detection of membrane proteins, but the improved sample preparation is still useful and enables detection of additional hydrophobic proteins. Proteolytic predigestion of plasma membranes enables detection of additional hydrophobic proteins and better sequence coverage of TMD-containing proteins in plasma membranes from both tissues.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

