Mechanistic basis of desmosome-targeted diseases
- PMID: 23911551
- PMCID: PMC3807649
- DOI: 10.1016/j.jmb.2013.07.035
Mechanistic basis of desmosome-targeted diseases
Abstract
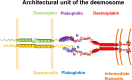
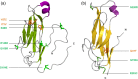
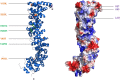
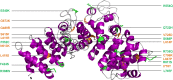
Desmosomes are dynamic junctions between cells that maintain the structural integrity of skin and heart tissues by withstanding shear forces. Mutations in component genes cause life-threatening conditions including arrhythmogenic right ventricular cardiomyopathy, and desmosomal proteins are targeted by pathogenic autoantibodies in skin blistering diseases such as pemphigus. Here, we review a set of newly discovered pathogenic alterations and discuss the structural repercussions of debilitating mutations on desmosomal proteins. The architectures of native desmosomal assemblies have been visualized by cryo-electron microscopy and cryo-electron tomography, and the network of protein domain interactions is becoming apparent. Plakophilin and desmoplakin mutations have been discovered to alter binding interfaces, structures, and stabilities of folded domains that have been resolved by X-ray crystallography and NMR spectroscopy. The flexibility within desmoplakin has been revealed by small-angle X-ray scattering and fluorescence assays, explaining how mechanical stresses are accommodated. These studies have shown that the structural and functional consequences of desmosomal mutations can now begin to be understood at multiple levels of spatial and temporal resolution. This review discusses the recent structural insights and raises the possibility of using modeling for mechanism-based diagnosis of how deleterious mutations alter the integrity of solid tissues.
Keywords: ARVC; DIFC; EC; EGFR; IA; ICS; Lef; NMD; PPAR; PRD; SH3; SR; Src homology 3; T-cell factor; Tcf; arrhythmogenic right ventricular cardiomyopathy; desmoplakin; desmosomal cadherin; desmosome; desmosome–intermediate filament complex; epidermal growth factor receptor; extracellular cadherin; iPSC; induced pluripotent stem cell; intracellular anchor; intracellular cadherin-typical sequence; lymphoid enhancer factor; nonsense-mediated RNA decay; peroxisome proliferator-activated receptor; plakin repeat domain; plakoglobin; spectrin repeat.
© 2013. Published by Elsevier Ltd. All rights reserved.
Figures


References
-
- Garrod D., Chidgey M. Desmosome structure, composition and function. Biochim Biophys Acta. 2008;1778:572–587. - PubMed
-
- Koeser J., Troyanovsky S.M., Grund C., Franke W.W. De novo formation of desmosomes in cultured cells upon transfection of genes encoding specific desmosomal components. Exp Cell Res. 2003;285:114–130. - PubMed
-
- Franke W.W., Borrmann C.M., Grund C., Pieperhoff S. The area composita of adhering junctions connecting heart muscle cells of vertebrates. I. Molecular definition in intercalated disks of cardiomyocytes by immunoelectron microscopy of desmosomal proteins. Eur J Cell Biol. 2006;85:69–82. - PubMed
-
- Borrmann C.M., Grund C., Kuhn C., Hofmann I., Pieperhoff S., Franke W.W. The area composita of adhering junctions connecting heart muscle cells of vertebrates. II. Colocalizations of desmosomal and fascia adhaerens molecules in the intercalated disk. Eur J Cell Biol. 2006;85:469–485. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

