p73 regulates autophagy and hepatocellular lipid metabolism through a transcriptional activation of the ATG5 gene
- PMID: 23912709
- PMCID: PMC3770317
- DOI: 10.1038/cdd.2013.104
p73 regulates autophagy and hepatocellular lipid metabolism through a transcriptional activation of the ATG5 gene
Abstract
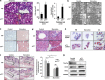
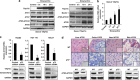
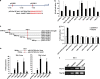
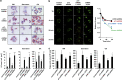
p73, a member of the p53 tumor suppressor family, is involved in neurogenesis, sensory pathways, immunity, inflammation, and tumorigenesis. How p73 is able to participate in such a broad spectrum of different biological processes is still largely unknown. Here, we report a novel role of p73 in regulating lipid metabolism by direct transactivation of the promoter of autophagy-related protein 5 (ATG5), a gene whose product is required for autophagosome formation. Following nutrient deprivation, the livers of p73-deficient mice demonstrate a massive accumulation of lipid droplets, together with a low level of autophagy, suggesting that triglyceride hydrolysis into fatty acids is blocked owing to deficient autophagy (macrolipophagy). Compared with wild-type mice, mice functionally deficient in all the p73 isoforms exhibit decreased ATG5 expression and lower levels of autophagy in multiple organs. We further show that the TAp73α is the critical p73 isoform responsible for inducing ATG5 expression in a p53-independent manner and demonstrate that ATG5 gene transfer can correct autophagy and macrolipophagy defects in p73-deficient hepatocytes. These data strongly suggest that the p73-ATG5 axis represents a novel, key pathway for regulating lipid metabolism through autophagy. The identification of p73 as a major regulator of autophagy suggests that it may have an important role in preventing or delaying disease and aging by maintaining a homeostatic control.
Figures

References
-
- Li Y, Prives C. Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function. Oncogene. 2007;26:2220–2225. - PubMed
-
- Levine AJ, Tomasini R, McKeon FD, Mak TW, Melino G. The p53 family: guardians of maternal reproduction. Nat Rev Mol Cell Biol. 2011;12:259–265. - PubMed
-
- Tomasini R, Mak TW, Melino G. The impact of p53 and p73 on aneuploidy and cancer. Trends Cell Biol. 2008;18:244–252. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

