MicroRNA-494 within an oncogenic microRNA megacluster regulates G1/S transition in liver tumorigenesis through suppression of mutated in colorectal cancer
- PMID: 23913442
- PMCID: PMC3877416
- DOI: 10.1002/hep.26662
MicroRNA-494 within an oncogenic microRNA megacluster regulates G1/S transition in liver tumorigenesis through suppression of mutated in colorectal cancer
Abstract
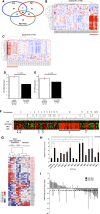
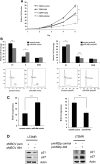
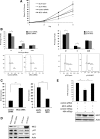
Hepatocellular carcinoma (HCC) is associated with poor survival for patients and few effective treatment options, raising the need for novel therapeutic strategies. MicroRNAs (miRNAs) play important roles in tumor development and show deregulated patterns of expression in HCC. Because of the liver's unique affinity for small nucleic acids, miRNA-based therapy has been proposed in the treatment of liver disease. Thus, there is an urgent need to identify and characterize aberrantly expressed miRNAs in HCC. In our study, we profiled miRNA expression changes in de novo liver tumors driven by MYC and/or RAS, two canonical oncogenes activated in a majority of human HCCs. We identified an up-regulated miRNA megacluster comprised of 53 miRNAs on mouse chromosome 12qF1 (human homolog 14q32). This miRNA megacluster is up-regulated in all three transgenic liver models and in a subset of human HCCs. An unbiased functional analysis of all miRNAs within this cluster was performed. We found that miR-494 is overexpressed in human HCC and aids in transformation by regulating the G1 /S cell cycle transition through targeting of the Mutated in Colorectal Cancer tumor suppressor. miR-494 inhibition in human HCC cell lines decreases cellular transformation, and anti-miR-494 treatment of primary MYC-driven liver tumor formation significantly diminishes tumor size.
Conclusion: Our findings identify a new therapeutic target (miR-494) for the treatment of HCC.
© 2013 Authors. Hepatology published by Wiley on behalf of the American Association for the Study of Liver Diseases.
Figures



References
-
- Bosch FX, Ribes J, Díaz M, Cléries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5–S16. - PubMed
-
- Schlaeger C, Longerich T, Schiller C, Bewerunge P, Mehrabi A, Toedt G, et al. Etiology-dependent molecular mechanisms in human hepatocarcinogenesis. Hepatology. 2008;47:511–520. - PubMed
-
- Gan FY, Gesell MS, Alousi M, Luk GD. Analysis of ODC and c-myc gene expression in hepatocellular carcinoma by in situ hybridization and immunohistochemistry. J Histochem Cytochem. 1993;41:1185–1196. - PubMed
-
- Calvisi DF, Ladu S, Gorden A, Farina M, Conner EA, Lee JS, et al. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology. 2006;130:1117–1128. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
