RNA-DNA differences in human mitochondria restore ancestral form of 16S ribosomal RNA
- PMID: 23913925
- PMCID: PMC3814879
- DOI: 10.1101/gr.161265.113
RNA-DNA differences in human mitochondria restore ancestral form of 16S ribosomal RNA
Abstract
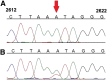
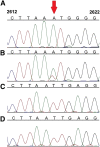
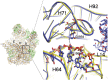
RNA transcripts are generally identical to the underlying DNA sequences. Nevertheless, RNA-DNA differences (RDDs) were found in the nuclear human genome and in plants and animals but not in human mitochondria. Here, by deep sequencing of human mitochondrial DNA (mtDNA) and RNA, we identified three RDD sites at mtDNA positions 295 (C-to-U), 13710 (A-to-U, A-to-G), and 2617 (A-to-U, A-to-G). Position 2617, within the 16S rRNA, harbored the most prevalent RDDs (>30% A-to-U and ∼15% A-to-G of the reads in all tested samples). The 2617 RDDs appeared already at the precursor polycistrone mitochondrial transcript. By using traditional Sanger sequencing, we identified the A-to-U RDD in six different cell lines and representative primates (Gorilla gorilla, Pongo pigmaeus, and Macaca mulatta), suggesting conservation of the mechanism generating such RDD. Phylogenetic analysis of more than 1700 vertebrate mtDNA sequences supported a thymine as the primate ancestral allele at position 2617, suggesting that the 2617 RDD recapitulates the ancestral 16S rRNA. Modeling U or G (the RDDs) at position 2617 stabilized the large ribosomal subunit structure in contrast to destabilization by an A (the pre-RDDs). Hence, these mitochondrial RDDs are likely functional.
Figures


References
-
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R 2013. Structures of the human and Drosophila 80S ribosome. Nature 497: 80–85 - PubMed
-
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M 2011. The structure of the eukaryotic ribosome at 3.0 A resolution. Science 334: 1524–1529 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
