Controls on bacterial and archaeal community structure and greenhouse gas production in natural, mined, and restored Canadian peatlands
- PMID: 23914185
- PMCID: PMC3728569
- DOI: 10.3389/fmicb.2013.00215
Controls on bacterial and archaeal community structure and greenhouse gas production in natural, mined, and restored Canadian peatlands
Abstract
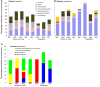
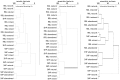
Northern peatlands are important global C reservoirs, largely because of their slow rates of microbial C mineralization. Particularly in sites that are heavily influenced by anthropogenic disturbances, there is scant information about microbial ecology and whether or not microbial community structure influences greenhouse gas production. This work characterized communities of bacteria and archaea using terminal restriction fragment length polymorphism (T-RFLP) and sequence analysis of 16S rRNA and functional genes across eight natural, mined, or restored peatlands in two locations in eastern Canada. Correlations were explored among chemical properties of peat, bacterial and archaeal community structure, and carbon dioxide (CO2) and methane (CH4) production rates under oxic and anoxic conditions. Bacteria and archaea similar to those found in other peat soil environments were detected. In contrast to other reports, methanogen diversity was low in our study, with only 2 groups of known or suspected methanogens. Although mining and restoration affected substrate availability and microbial activity, these land-uses did not consistently affect bacterial or archaeal community composition. In fact, larger differences were observed between the two locations and between oxic and anoxic peat samples than between natural, mined, and restored sites, with anoxic samples characterized by less detectable bacterial diversity and stronger dominance by members of the phylum Acidobacteria. There were also no apparent strong linkages between prokaryote community structure and CH4 or CO2 production, suggesting that different organisms exhibit functional redundancy and/or that the same taxa function at very different rates when exposed to different peat substrates. In contrast to other earlier work focusing on fungal communities across similar mined and restored peatlands, bacterial and archaeal communities appeared to be more resistant or resilient to peat substrate changes brought about by these land uses.
Keywords: archaea; bacteria; carbon dioxide; decomposition; methane; methanogen.
Figures



References
-
- Andersen R., Chapman S. J., Artz R. R. E. (2013). Microbial communities in natural and disturbed peatlands: a review. Soil Biol. Biochem. 57, 979–994 10.1016/j.soilbio.2012.10.003 - DOI
-
- Andersen R., Francez A. J., Rochefort L. (2006). The physicochemical and microbiological status of a restored bog in Quebéc: identification of relevant criteria to monitor success. Soil Biol. Biochem. 38, 1375–1387 10.1016/j.soilbio.2005.10.012 - DOI
-
- Artz R. R. E., Chapman S. J., Siegenthaler A., Mitchell E. A. D., Buttler A., Bortoluzzi E., et al. (2008). Functional microbial diversity in regenerating cutover peatlands responds to vegetation succession. Appl. Soil Ecol. 45, 1799–1809 10.1111/j.1365-2664.2008.01573.x - DOI
-
- Ausec L., Kraigher B., Mandic-Mulec I. (2009). Differences in the activity and bacterial community structure of drained grassland and forest peat soils. Soil Biol. Biochem. 41, 1874–1881 10.1016/j.soilbio.2009.06.010 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

