Proteomic characterization of novel histone post-translational modifications
- PMID: 23916056
- PMCID: PMC3737111
- DOI: 10.1186/1756-8935-6-24
Proteomic characterization of novel histone post-translational modifications
Abstract
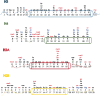
Histone post-translational modifications (PTMs) have been linked to a variety of biological processes and disease states, thus making their characterization a critical field of study. In the last 5 years, a number of novel sites and types of modifications have been discovered, greatly expanding the histone code. Mass spectrometric methods are essential for finding and validating histone PTMs. Additionally, novel proteomic, genomic and chemical biology tools have been developed to probe PTM function. In this snapshot review, proteomic tools for PTM identification and characterization will be discussed and an overview of PTMs found in the last 5 years will be provided.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

