DNA methylation and differentiation: HOX genes in muscle cells
- PMID: 23916067
- PMCID: PMC3750649
- DOI: 10.1186/1756-8935-6-25
DNA methylation and differentiation: HOX genes in muscle cells
Abstract
Background: Tight regulation of homeobox genes is essential for vertebrate development. In a study of genome-wide differential methylation, we recently found that homeobox genes, including those in the HOX gene clusters, were highly overrepresented among the genes with hypermethylation in the skeletal muscle lineage. Methylation was analyzed by reduced representation bisulfite sequencing (RRBS) of postnatal myoblasts, myotubes and adult skeletal muscle tissue and 30 types of non-muscle-cell cultures or tissues.
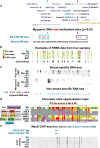
Results: In this study, we found that myogenic hypermethylation was present in specific subregions of all four HOX gene clusters and was associated with various chromatin epigenetic features. Although the 3' half of the HOXD cluster was silenced and enriched in polycomb repression-associated H3 lysine 27 trimethylation in most examined cell types, including myoblasts and myotubes, myogenic samples were unusual in also displaying much DNA methylation in this region. In contrast, both HOXA and HOXC clusters displayed myogenic hypermethylation bordering a central region containing many genes preferentially expressed in myogenic progenitor cells and consisting largely of chromatin with modifications typical of promoters and enhancers in these cells. A particularly interesting example of myogenic hypermethylation was HOTAIR, a HOXC noncoding RNA gene, which can silence HOXD genes in trans via recruitment of polycomb proteins. In myogenic progenitor cells, the preferential expression of HOTAIR was associated with hypermethylation immediately downstream of the gene. Other HOX gene regions also displayed myogenic DNA hypermethylation despite being moderately expressed in myogenic cells. Analysis of representative myogenic hypermethylated sites for 5-hydroxymethylcytosine revealed little or none of this base, except for an intragenic site in HOXB5 which was specifically enriched in this base in skeletal muscle tissue, whereas myoblasts had predominantly 5-methylcytosine at the same CpG site.
Conclusions: Our results suggest that myogenic hypermethylation of HOX genes helps fine-tune HOX sense and antisense gene expression through effects on 5' promoters, intragenic and intergenic enhancers and internal promoters. Myogenic hypermethylation might also affect the relative abundance of different RNA isoforms, facilitate transcription termination, help stop the spread of activation-associated chromatin domains and stabilize repressive chromatin structures.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

