Molecular basis of HHQ biosynthesis: molecular dynamics simulations, enzyme kinetic and surface plasmon resonance studies
- PMID: 23916145
- PMCID: PMC3734052
- DOI: 10.1186/2046-1682-6-10
Molecular basis of HHQ biosynthesis: molecular dynamics simulations, enzyme kinetic and surface plasmon resonance studies
Abstract
Background: PQS (PseudomonasQuinolone Signal) and its precursor HHQ are signal molecules of the P. aeruginosa quorum sensing system. They explicate their role in mammalian pathogenicity by binding to the receptor PqsR that induces virulence factor production and biofilm formation. The enzyme PqsD catalyses the biosynthesis of HHQ.
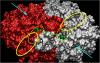
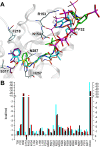
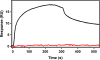
Results: Enzyme kinetic analysis and surface plasmon resonance (SPR) biosensor experiments were used to determine mechanism and substrate order of the biosynthesis. Comparative analysis led to the identification of domains involved in functionality of PqsD. A kinetic cycle was set up and molecular dynamics (MD) simulations were used to study the molecular bases of the kinetics of PqsD. Trajectory analysis, pocket volume measurements, binding energy estimations and decompositions ensured insights into the binding mode of the substrates anthraniloyl-CoA and β-ketodecanoic acid.
Conclusions: Enzyme kinetics and SPR experiments hint at a ping-pong mechanism for PqsD with ACoA as first substrate. Trajectory analysis of different PqsD complexes evidenced ligand-dependent induced-fit motions affecting the modified ACoA funnel access to the exposure of a secondary channel. A tunnel-network is formed in which Ser317 plays an important role by binding to both substrates. Mutagenesis experiments resulting in the inactive S317F mutant confirmed the importance of this residue. Two binding modes for β-ketodecanoic acid were identified with distinct catalytic mechanism preferences.
Figures



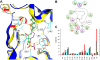


Similar articles
-
Mechanistic details for anthraniloyl transfer in PqsD: the initial step in HHQ biosynthesis.J Mol Model. 2014 Jun;20(6):2255. doi: 10.1007/s00894-014-2255-z. Epub 2014 May 15. J Mol Model. 2014. PMID: 24842325
-
Structure optimization of 2-benzamidobenzoic acids as PqsD inhibitors for Pseudomonas aeruginosa infections and elucidation of binding mode by SPR, STD NMR, and molecular docking.J Med Chem. 2013 Aug 8;56(15):6146-55. doi: 10.1021/jm4006302. Epub 2013 Jul 19. J Med Chem. 2013. PMID: 23834469
-
Catechol-based substrates of chalcone synthase as a scaffold for novel inhibitors of PqsD.Eur J Med Chem. 2015 Jan 27;90:351-9. doi: 10.1016/j.ejmech.2014.11.055. Epub 2014 Nov 27. Eur J Med Chem. 2015. PMID: 25437621
-
Recent Advances in the Discovery of PqsD Inhibitors as Antimicrobial Agents.ChemMedChem. 2017 Mar 17;12(6):420-425. doi: 10.1002/cmdc.201700015. Epub 2017 Mar 6. ChemMedChem. 2017. PMID: 28195681 Review.
-
Anti-PqsR compounds as next-generation antibacterial agents against Pseudomonas aeruginosa: A review.Eur J Med Chem. 2019 Jun 15;172:26-35. doi: 10.1016/j.ejmech.2019.03.049. Epub 2019 Mar 25. Eur J Med Chem. 2019. PMID: 30939351 Review.
Cited by
-
PqsBC, a Condensing Enzyme in the Biosynthesis of the Pseudomonas aeruginosa Quinolone Signal: CRYSTAL STRUCTURE, INHIBITION, AND REACTION MECHANISM.J Biol Chem. 2016 Mar 25;291(13):6610-24. doi: 10.1074/jbc.M115.708453. Epub 2016 Jan 25. J Biol Chem. 2016. PMID: 26811339 Free PMC article.
-
Synthesis, In Silico, and In Vitro Evaluation of Long Chain Alkyl Amides from 2-Amino-4-Quinolone Derivatives as Biofilm Inhibitors.Molecules. 2019 Jan 17;24(2):327. doi: 10.3390/molecules24020327. Molecules. 2019. PMID: 30658415 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

