Molecular architecture of the mammalian circadian clock
- PMID: 23916625
- PMCID: PMC3946763
- DOI: 10.1016/j.tcb.2013.07.002
Molecular architecture of the mammalian circadian clock
Abstract
Circadian clocks coordinate physiology and behavior with the 24h solar day to provide temporal homeostasis with the external environment. The molecular clocks that drive these intrinsic rhythmic changes are based on interlocked transcription/translation feedback loops that integrate with diverse environmental and metabolic stimuli to generate internal 24h timing. In this review we highlight recent advances in our understanding of the core molecular clock and how it utilizes diverse transcriptional and post-transcriptional mechanisms to impart temporal control onto mammalian physiology. Understanding the way in which biological rhythms are generated throughout the body may provide avenues for temporally directed therapeutics to improve health and prevent disease.
Keywords: circadian; peripheral clock; post-transcription; transcription.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures

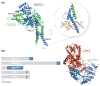
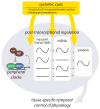
References
-
- Ko C, Takahashi J. Molecular components of the mammalian circadian clock. Human molecular genetics. 2006;15(Spec No 2):7. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

