Two mammalian MAGOH genes contribute to exon junction complex composition and nonsense-mediated decay
- PMID: 23917022
- PMCID: PMC3817150
- DOI: 10.4161/rna.25827
Two mammalian MAGOH genes contribute to exon junction complex composition and nonsense-mediated decay
Abstract
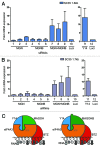
The exon junction complex (EJC) participates in the regulation of many post-transcriptional steps of gene expression. EJCs are deposited on messenger RNAs (mRNAs) during splicing and their core consists of eIF4A3, MLN51, Y14, and MAGOH. Here, we show that two genes encoding MAGOH paralogs (referred to as MAGOH and MAGOHB) are expressed in mammals. In macrophages, the expression of MAGOHB, but not MAGOH mRNA, increases rapidly after LPS stimulation. Both MAGOH proteins interact with other EJC components, incorporate into mRNA-bound EJCs, and activate nonsense-mediated decay. Furthermore, the simultaneous depletion of MAGOH and MAGOHB, but not individual depletions, impair nonsense-mediated decay in human cells. Hence, our results establish that the core composition of mammalian EJCs is more complex than previously recognized.
Keywords: EJC; MAGOHB; NMD; gene duplication; mago nashi homolog.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
