A systematic analysis of the PARP protein family identifies new functions critical for cell physiology
- PMID: 23917125
- PMCID: PMC3756671
- DOI: 10.1038/ncomms3240
A systematic analysis of the PARP protein family identifies new functions critical for cell physiology
Abstract
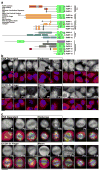
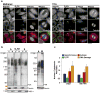
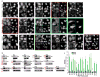
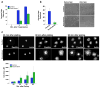
The poly(ADP-ribose) polymerase (PARP) family of proteins use NAD(+) as their substrate to modify acceptor proteins with ADP-ribose modifications. The function of most PARPs under physiological conditions is unknown. Here, to better understand this protein family, we systematically analyse the cell cycle localization of each PARP and of poly(ADP-ribose), a product of PARP activity, then identify the knockdown phenotype of each protein and perform secondary assays to elucidate function. We show that most PARPs are cytoplasmic, identify cell cycle differences in the ratio of nuclear to cytoplasmic poly(ADP-ribose) and identify four phenotypic classes of PARP function. These include the regulation of membrane structures, cell viability, cell division and the actin cytoskeleton. Further analysis of PARP14 shows that it is a component of focal adhesion complexes required for proper cell motility and focal adhesion function. In total, we show that PARP proteins are critical regulators of eukaryotic physiology.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21:255–261. - PubMed
-
- Bürkle A. Poly(ADP-ribose): The most elaborate metabolite of NAD+ FEBS J. 2005;272:4576–4589. - PubMed
-
- Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem Cell Biol. 2005;83:354–364. - PubMed
-
- Koh DW, Dawson TM, Dawson VL. Mediation of cell death by poly(ADP-ribose) polymerase-1. Pharmacol Res. 2005;52:5–14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

