Association of maternal and nutrient supply line factors with DNA methylation at the imprinted IGF2/H19 locus in multiple tissues of newborn twins
- PMID: 23917818
- PMCID: PMC3891688
- DOI: 10.4161/epi.25908
Association of maternal and nutrient supply line factors with DNA methylation at the imprinted IGF2/H19 locus in multiple tissues of newborn twins
Abstract
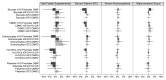
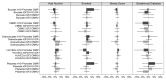
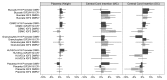
Epigenetic events are crucial for early development, but can be influenced by environmental factors, potentially programming the genome for later adverse health outcomes. The insulin-like growth factor 2 (IGF2)/H19 locus is crucial for prenatal growth and the epigenetic state at this locus is environmentally labile. Recent studies have implicated maternal factors, including folate intake and smoking, in the regulation of DNA methylation at this locus, although data are often conflicting in the direction and magnitude of effect. Most studies have focused on single tissues and on one or two differentially-methylated regions (DMRs) regulating IGF2/H19 expression. In this study, we investigated the relationship between multiple shared and non-shared gestational/maternal factors and DNA methylation at four IGF2/H19 DMRs in five newborn cell types from 67 pairs of monozygotic and 49 pairs of dizygotic twins. Data on maternal and non-shared supply line factors were collected during the second and third trimesters of pregnancy and DNA methylation was measured via mass spectrometry using Sequenom MassArray EpiTyper analysis. Our exploratory approach showed that the site of umbilical cord insertion into the placenta in monochorionic twins has the strongest positive association with methylation in all IGF2/H19 DMRs (p<0.05). Further, evidence for tissue- and locus-specific effects were observed, emphasizing that responsiveness to environmental exposures in utero cannot be generalized across genes and tissues, potentially accounting for the lack of consistency in previous findings. Such complexity in responsiveness to environmental exposures in utero has implications for all epigenetic studies investigating the developmental origins of health and disease.
Keywords: DNA methylation; developmental origins of health and disease (DOHaD); imprinted genes; maternal factors; twins.
Figures
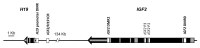
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
