ETV4 promotes metastasis in response to activation of PI3-kinase and Ras signaling in a mouse model of advanced prostate cancer
- PMID: 23918374
- PMCID: PMC3773788
- DOI: 10.1073/pnas.1303558110
ETV4 promotes metastasis in response to activation of PI3-kinase and Ras signaling in a mouse model of advanced prostate cancer
Abstract
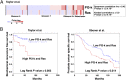
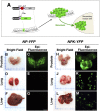
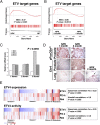
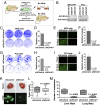
Combinatorial activation of PI3-kinase and RAS signaling occurs frequently in advanced prostate cancer and is associated with adverse patient outcome. We now report that the oncogenic Ets variant 4 (Etv4) promotes prostate cancer metastasis in response to coactivation of PI3-kinase and Ras signaling pathways in a genetically engineered mouse model of highly penetrant, metastatic prostate cancer. Using an inducible Cre driver to simultaneously inactivate Pten while activating oncogenic Kras and a fluorescent reporter allele in the prostate epithelium, we performed lineage tracing in vivo to define the temporal and spatial occurrence of prostate tumors, disseminated tumor cells, and metastases. These analyses revealed that though disseminated tumors cells arise early following the initial occurrence of prostate tumors, there is a significant temporal lag in metastasis, which is temporally coincident with the up-regulation of Etv4 expression in primary tumors. Functional studies showed that knockdown of Etv4 in a metastatic cell line derived from the mouse model abrogates the metastatic phenotype but does not affect tumor growth. Notably, expression and activation of ETV4, but not other oncogenic ETS genes, is correlated with activation of both PI3-kinase and Ras signaling in human prostate tumors and metastases. Our findings indicate that ETV4 promotes metastasis in prostate tumors that have activation of PI3-kinase and Ras signaling, and therefore, ETV4 represents a potential target of therapeutic intervention for metastatic prostate cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Comment in
-
Understanding the temporal sequence of genetic events that lead to prostate cancer progression and metastasis.Proc Natl Acad Sci U S A. 2013 Sep 10;110(37):14819-20. doi: 10.1073/pnas.1313997110. Epub 2013 Aug 30. Proc Natl Acad Sci U S A. 2013. PMID: 23995446 Free PMC article. No abstract available.
References
-
- Nguyen DX, Bos PD, Massagué J. Metastasis: From dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9(4):274–284. - PubMed
-
- American Cancer Society (2012) Cancer Facts and Figures 2012. Available at www.cancer.org/acs/groups/content/@epidemiologysurveilance/documents/acs.... Accessed September 25, 2012.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

