Epigenetic regulation of autophagy by the methyltransferase G9a
- PMID: 23918802
- PMCID: PMC3811684
- DOI: 10.1128/MCB.00813-13
Epigenetic regulation of autophagy by the methyltransferase G9a
Abstract
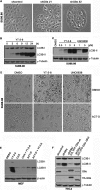
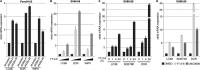
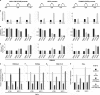
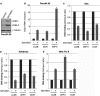
Macroautophagy is an evolutionarily conserved cellular process involved in the clearance of proteins and organelles. Although the cytoplasmic machinery that orchestrates autophagy induction during starvation, hypoxia, or receptor stimulation has been widely studied, the key epigenetic events that initiate and maintain the autophagy process remain unknown. Here we show that the methyltransferase G9a coordinates the transcriptional activation of key regulators of autophagosome formation by remodeling the chromatin landscape. Pharmacological inhibition or RNA interference (RNAi)-mediated suppression of G9a induces LC3B expression and lipidation that is dependent on RNA synthesis, protein translation, and the methyltransferase activity of G9a. Under normal conditions, G9a associates with the LC3B, WIPI1, and DOR gene promoters, epigenetically repressing them. However, G9a and G9a-repressive histone marks are removed during starvation and receptor-stimulated activation of naive T cells, two physiological inducers of macroautophagy. Moreover, we show that the c-Jun N-terminal kinase (JNK) pathway is involved in the regulation of autophagy gene expression during naive-T-cell activation. Together, these findings reveal that G9a directly represses genes known to participate in the autophagic process and that inhibition of G9a-mediated epigenetic repression represents an important regulatory mechanism during autophagy.
Figures



Comment in
-
Histone methylation keeps the brakes on autophagy.Mol Cell Biol. 2013 Oct;33(20):3974-5. doi: 10.1128/MCB.01033-13. Epub 2013 Aug 26. Mol Cell Biol. 2013. PMID: 23979595 Free PMC article. No abstract available.
References
-
- Klionsky DJ. 2007. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8:931–937 - PubMed
-
- Mehrpour M, Esclatine A, Beau I, Codogno P. 2010. Overview of macroautophagy regulation in mammalian cells. Cell Res. 20:748–762 - PubMed
-
- Choi AMK, Ryter SW, Levine B. 2013. Autophagy in human health and disease. N. Engl. J. Med. 368:651–662 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
