The switch-associated protein 70 (SWAP-70) bundles actin filaments and contributes to the regulation of F-actin dynamics
- PMID: 23921380
- PMCID: PMC3789966
- DOI: 10.1074/jbc.M113.461277
The switch-associated protein 70 (SWAP-70) bundles actin filaments and contributes to the regulation of F-actin dynamics
Abstract
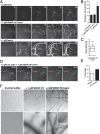
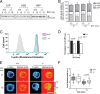
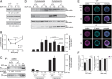
Coordinated assembly and disassembly of actin into filaments and higher order structures such as stress fibers and lamellipodia are fundamental for cell migration and adhesion. However, the precise spatiotemporal regulation of F-actin structures is not completely understood. SWAP-70, a phosphatidylinositol 3,4,5-trisphosphate-interacting, F-actin-binding protein, participates in actin rearrangements through yet unknown mechanisms. Here, we show that SWAP-70 is an F-actin-bundling protein that oligomerizes through a Gln/Glu-rich stretch within a coiled-coil region. SWAP-70 bundles filaments in parallel and anti-parallel fashion through its C-terminal F-actin binding domain and delays dilution-induced F-actin depolymerization. We further demonstrate that SWAP-70 co-localizes and directly interacts with cofilin, an F-actin severing and depolymerization factor, and contributes to the regulation of cofilin activity in vivo. In line with these activities, upon stem cell factor stimulation, murine bone marrow-derived mast cells lacking SWAP-70 display aberrant regulation of F-actin and actin free barbed ends dynamics. Moreover, proper stem cell factor-dependent cofilin activation via dephosphorylation and subcellular redistribution into a detergent-resistant cytoskeletal compartment also require SWAP-70. Together, these findings reveal an important role of SWAP-70 in the dynamic spatiotemporal regulation of F-actin networks.
Keywords: Actin; Cell Biology; Cofilin; Cytoskeleton; F-actin Bundling; Mast Cell; SWAP-70.
Figures





Similar articles
-
The F-actin bundler SWAP-70 promotes tumor metastasis.Life Sci Alliance. 2024 May 17;7(8):e202302307. doi: 10.26508/lsa.202302307. Print 2024 Aug. Life Sci Alliance. 2024. PMID: 38760173 Free PMC article.
-
SWAP-70 regulates c-kit-induced mast cell activation, cell-cell adhesion, and migration.Mol Cell Biol. 2004 Dec;24(23):10277-88. doi: 10.1128/MCB.24.23.10277-10288.2004. Mol Cell Biol. 2004. PMID: 15542837 Free PMC article.
-
Mechanism of control of F-actin cortex architecture by SWAP-70.J Cell Sci. 2020 Jan 23;133(2):jcs233064. doi: 10.1242/jcs.233064. J Cell Sci. 2020. PMID: 31932501
-
Functions of actin-interacting protein 1 (AIP1)/WD repeat protein 1 (WDR1) in actin filament dynamics and cytoskeletal regulation.Biochem Biophys Res Commun. 2018 Nov 25;506(2):315-322. doi: 10.1016/j.bbrc.2017.10.096. Epub 2017 Oct 19. Biochem Biophys Res Commun. 2018. PMID: 29056508 Free PMC article. Review.
-
Juxtanuclear Drebrin-Enriched Zone.Adv Exp Med Biol. 2017;1006:329-336. doi: 10.1007/978-4-431-56550-5_19. Adv Exp Med Biol. 2017. PMID: 28865029 Review.
Cited by
-
The F-actin modulator SWAP-70 controls podosome patterning in osteoclasts.Bone Rep. 2016 Jul 19;5:214-221. doi: 10.1016/j.bonr.2016.07.002. eCollection 2016 Dec. Bone Rep. 2016. PMID: 28580389 Free PMC article.
-
SWAP70 Organizes the Actin Cytoskeleton and Is Essential for Phagocytosis.Cell Rep. 2016 Nov 1;17(6):1518-1531. doi: 10.1016/j.celrep.2016.10.021. Cell Rep. 2016. PMID: 27806292 Free PMC article.
-
The Actin Cytoskeleton at the Immunological Synapse of Dendritic Cells.Front Cell Dev Biol. 2021 Aug 2;9:679500. doi: 10.3389/fcell.2021.679500. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34409027 Free PMC article. Review.
-
RhoA Signaling in Immune Cell Response and Cardiac Disease.Cells. 2021 Jul 3;10(7):1681. doi: 10.3390/cells10071681. Cells. 2021. PMID: 34359851 Free PMC article. Review.
-
The guanine exchange factor SWAP70 mediates vGPCR-induced endothelial plasticity.Cell Commun Signal. 2015 Feb 15;13:11. doi: 10.1186/s12964-015-0090-1. Cell Commun Signal. 2015. PMID: 25889342 Free PMC article.
References
-
- Pollard T. D., Borisy G. G. (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465 - PubMed
-
- Le Clainche C., Carlier M. F. (2008) Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol. Rev. 88, 489–513 - PubMed
-
- Stevenson R. P., Veltman D., Machesky L. M. (2012) Actin-bundling proteins in cancer progression at a glance. J. Cell Sci. 125, 1073–1079 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

