The Bin/amphiphysin/Rvs (BAR) domain protein endophilin B2 interacts with plectin and controls perinuclear cytoskeletal architecture
- PMID: 23921385
- PMCID: PMC3779758
- DOI: 10.1074/jbc.M113.485482
The Bin/amphiphysin/Rvs (BAR) domain protein endophilin B2 interacts with plectin and controls perinuclear cytoskeletal architecture
Abstract
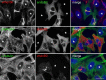
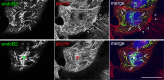
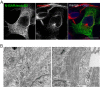
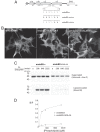
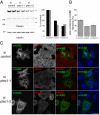
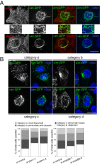
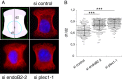
Proteins of the Bin/amphiphysin/Rvs (BAR) domain superfamily are essential in controlling the shape and dynamics of intracellular membranes. Here, we present evidence for the unconventional function of a member of the endophilin family of BAR and Src homology 3 domain-containing proteins, namely endophilin B2, in the perinuclear organization of intermediate filaments. Using mass spectrometry analysis based on capturing endophilin B2 partners in in situ pre-established complexes in cells, we unravel the interaction of endophilin B2 with plectin 1, a variant of the cytoskeleton linker protein plectin as well as with vimentin. Endophilin B2 directly binds the N-terminal region of plectin 1 via Src homology 3-mediated interaction and vimentin indirectly via plectin-mediated interaction. The relevance of these interactions is strengthened by the selective and drastic reorganization of vimentin around nuclei upon overexpression of endophilin B2 and by the extensive colocalization of both proteins in a meshwork of perinuclear filamentous structures. By generating mutants of the endophilin B2 BAR domain, we show that this phenotype requires the BAR-mediated membrane binding activity of endophilin B2. Plectin 1 or endophilin B2 knockdown using RNA interference disturbed the perinuclear organization of vimentin. Altogether, these data suggest that the endophilin B2-plectin 1 complex functions as a membrane-anchoring device organizing and stabilizing the perinuclear network of vimentin filaments. Finally, we present evidence for the involvement of endophilin B2 and plectin 1 in nuclear positioning in individual cells. This points to the potential importance of the endophilin B2-plectin complex in the biological functions depending on nuclear migration and positioning.
Keywords: BAR Domain; Cell Biology; Cytoskeleton; Endophilin; Intermediate Filaments; Membrane; Plectin; Protein Assembly; SH3 Domains.
Figures




References
-
- McMahon H. T., Gallop J. L. (2005) Membrane curvature and mechanisms of dynamic cell membrane remodeling. Nature 438, 590–596 - PubMed
-
- Itoh T., De Camilli P. (2006) BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim. Biophys. Acta 1761, 897–912 - PubMed
-
- Zimmerberg J., Kozlov M. M. (2006) How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 7, 9–19 - PubMed
-
- Masuda M., Mochizuki N. (2010) Structural characteristics of BAR domain superfamily to sculpt the membrane. Semin. Cell Dev. Biol. 21, 391–398 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

