Transcription activator like effector (TALE)-directed piggyBac transposition in human cells
- PMID: 23921635
- PMCID: PMC3799441
- DOI: 10.1093/nar/gkt677
Transcription activator like effector (TALE)-directed piggyBac transposition in human cells
Abstract
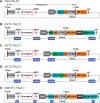
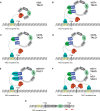
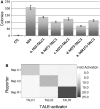
Insertional therapies have shown great potential for combating genetic disease and safer methods would undoubtedly broaden the variety of possible illness that can be treated. A major challenge that remains is reducing the risk of insertional mutagenesis due to random insertion by both viral and non-viral vectors. Targetable nucleases are capable of inducing double-stranded breaks to enhance homologous recombination for the introduction of transgenes at specific sequences. However, off-target DNA cleavages at unknown sites can lead to mutations that are difficult to detect. Alternatively, the piggyBac transposase is able perform all of the steps required for integration; therefore, cells confirmed to contain a single copy of a targeted transposon, for which its location is known, are likely to be devoid of aberrant genomic modifications. We aimed to retarget transposon insertions by comparing a series of novel hyperactive piggyBac constructs tethered to a custom transcription activator like effector DNA-binding domain designed to bind the first intron of the human CCR5 gene. Multiple targeting strategies were evaluated using combinations of both plasmid-DNA and transposase-protein relocalization to the target sequence. We demonstrated user-defined directed transposition to the CCR5 genomic safe harbor and isolated single-copy clones harboring targeted integrations.
Figures


Similar articles
-
Vast potential for using the piggyBac transposon to engineer transgenic plants at specific genomic locations.Bioengineered. 2016;7(1):3-6. doi: 10.1080/21655979.2015.1131367. Bioengineered. 2016. PMID: 26930269 Free PMC article.
-
Comparative analysis of chimeric ZFP-, TALE- and Cas9-piggyBac transposases for integration into a single locus in human cells.Nucleic Acids Res. 2017 Aug 21;45(14):8411-8422. doi: 10.1093/nar/gkx572. Nucleic Acids Res. 2017. PMID: 28666380 Free PMC article.
-
Chimeric piggyBac transposases for genomic targeting in human cells.Nucleic Acids Res. 2012 Aug;40(14):6978-91. doi: 10.1093/nar/gks309. Epub 2012 Apr 9. Nucleic Acids Res. 2012. PMID: 22492708 Free PMC article.
-
Transposase concentration controls transposition activity: myth or reality?Gene. 2013 Nov 10;530(2):165-71. doi: 10.1016/j.gene.2013.08.039. Epub 2013 Aug 28. Gene. 2013. PMID: 23994686 Review.
-
Contemporary Transposon Tools: A Review and Guide through Mechanisms and Applications of Sleeping Beauty, piggyBac and Tol2 for Genome Engineering.Int J Mol Sci. 2021 May 11;22(10):5084. doi: 10.3390/ijms22105084. Int J Mol Sci. 2021. PMID: 34064900 Free PMC article. Review.
Cited by
-
Evaluating different DNA binding domains to modulate L1 ORF2p-driven site-specific retrotransposition events in human cells.Gene. 2018 Feb 5;642:188-198. doi: 10.1016/j.gene.2017.11.033. Epub 2017 Nov 14. Gene. 2018. PMID: 29154869 Free PMC article.
-
Optimized tuning of TALEN specificity using non-conventional RVDs.Sci Rep. 2015 Jan 30;5:8150. doi: 10.1038/srep08150. Sci Rep. 2015. PMID: 25632877 Free PMC article.
-
An Engineered Cas-Transposon System for Programmable and Site-Directed DNA Transpositions.CRISPR J. 2019 Dec;2(6):376-394. doi: 10.1089/crispr.2019.0030. Epub 2019 Nov 19. CRISPR J. 2019. PMID: 31742433 Free PMC article.
-
Precise genetic engineering with piggyBac transposon in plants.Plant Biotechnol (Tokyo). 2023 Dec 25;40(4):255-262. doi: 10.5511/plantbiotechnology.23.0525a. Plant Biotechnol (Tokyo). 2023. PMID: 38434112 Free PMC article.
-
Vast potential for using the piggyBac transposon to engineer transgenic plants at specific genomic locations.Bioengineered. 2016;7(1):3-6. doi: 10.1080/21655979.2015.1131367. Bioengineered. 2016. PMID: 26930269 Free PMC article.

