Chromatin loop organization of the junb locus in mouse dendritic cells
- PMID: 23921639
- PMCID: PMC3799436
- DOI: 10.1093/nar/gkt669
Chromatin loop organization of the junb locus in mouse dendritic cells
Abstract
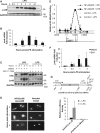
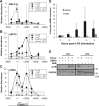
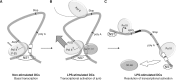
The junb gene behaves as an immediate early gene in bacterial lipopolysaccharide (LPS)-stimulated dendritic cells (DCs), where its transient transcriptional activation is necessary for the induction of inflammatory cytokines. junb is a short gene and its transcriptional activation by LPS depends on the binding of NF-κB to an enhancer located just downstream of its 3' UTR. Here, we have addressed the mechanisms underlying the transcriptional hyper-reactivity of junb. Using transfection and pharmacological assays to complement chromatin immunoprecipitation analyses addressing the localization of histones, polymerase II, negative elongation factor (NELF)-, DRB sensitivity-inducing factor (DSIF)- and Positive Transcription Factor b complexes, we demonstrate that junb is a RNA Pol II-paused gene where Pol II is loaded in the transcription start site domain but poorly active. Moreover, High salt-Recovered Sequence, chromosome conformation capture (3C)- and gene transfer experiments show that (i) junb is organized in a nuclear chromatin loop bringing into close spatial proximity the upstream promoter region and the downstream enhancer and (ii) this configuration permits immediate Pol II release on the junb body on binding of LPS-activated NF-κB to the enhancer. Thus, our work unveils a novel topological framework underlying fast junb transcriptional response in DCs. Moreover, it also points to a novel layer of complexity in the modes of action of NF-κB.
Figures






Similar articles
-
An NF-kappaB-dependent role for JunB in the induction of proinflammatory cytokines in LPS-activated bone marrow-derived dendritic cells.PLoS One. 2010 Mar 8;5(3):e9585. doi: 10.1371/journal.pone.0009585. PLoS One. 2010. PMID: 20221401 Free PMC article.
-
Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene.Mol Cell Biol. 2006 Aug;26(16):6094-104. doi: 10.1128/MCB.02366-05. Mol Cell Biol. 2006. PMID: 16880520 Free PMC article.
-
NF-kappaB elements contribute to junB inducibility by lipopolysaccharide in the murine macrophage cell line RAW264.7.FEBS Lett. 2002 Feb 27;513(2-3):203-7. doi: 10.1016/s0014-5793(02)02295-0. FEBS Lett. 2002. PMID: 11904151
-
Emerging roles of transcriptional enhancers in chromatin looping and promoter-proximal pausing of RNA polymerase II.J Biol Chem. 2018 Sep 7;293(36):13786-13794. doi: 10.1074/jbc.R117.813485. Epub 2017 Nov 29. J Biol Chem. 2018. PMID: 29187597 Free PMC article. Review.
-
Regulation of the MIR155 host gene in physiological and pathological processes.Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review.
Cited by
-
Novel insights into chromosomal conformations in cancer.Mol Cancer. 2017 Nov 17;16(1):173. doi: 10.1186/s12943-017-0741-5. Mol Cancer. 2017. PMID: 29149895 Free PMC article. Review.
-
Contribution of Topological Domains and Loop Formation to 3D Chromatin Organization.Genes (Basel). 2015 Jul 27;6(3):734-50. doi: 10.3390/genes6030734. Genes (Basel). 2015. PMID: 26226004 Free PMC article. Review.
-
Gene-to-gene coordinated regulation of transcription and alternative splicing by 3D chromatin remodeling upon NF-κB activation.Nucleic Acids Res. 2024 Feb 28;52(4):1527-1543. doi: 10.1093/nar/gkae015. Nucleic Acids Res. 2024. PMID: 38272542 Free PMC article.
-
JunB: a paradigm for Jun family in immune response and cancer.Front Cell Infect Microbiol. 2023 Sep 4;13:1222265. doi: 10.3389/fcimb.2023.1222265. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37731821 Free PMC article. Review.
-
Constitutive Activation of NF-κB Pathway in Hematopoietic Stem Cells Causes Loss of Quiescence and Deregulated Transcription Factor Networks.Front Cell Dev Biol. 2018 Oct 30;6:143. doi: 10.3389/fcell.2018.00143. eCollection 2018. Front Cell Dev Biol. 2018. PMID: 30425986 Free PMC article.
References
-
- Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, Pulendran B, Palucka K. Immunobiology of dendritic cells. Annu. Rev. Immunol. 2000;18:767–811. - PubMed
-
- Heath WR, Carbone FR. Dendritic cell subsets in primary and secondary T cell responses at body surfaces. Nat. Immunol. 2009;10:1237–1244. - PubMed
-
- Reis e Sousa C. Dendritic cells in a mature age. Nat. Rev. Immunol. 2006;6:476–483. - PubMed
-
- Steinman RM, Banchereau J. Taking dendritic cells into medicine. Nature. 2007;449:419–426. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

