Diagnostic applications of next generation sequencing in immunogenetics and molecular oncology
- PMID: 23922545
- PMCID: PMC3725031
- DOI: 10.1159/000351267
Diagnostic applications of next generation sequencing in immunogenetics and molecular oncology
Abstract
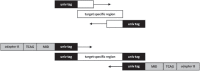
With the introduction of the next generation sequencing (NGS) technologies, remarkable new diagnostic applications have been established in daily routine. Implementation of NGS is challenging in clinical diagnostics, but definite advantages and new diagnostic possibilities make the switch to the technology inevitable. In addition to the higher sequencing capacity, clonal sequencing of single molecules, multiplexing of samples, higher diagnostic sensitivity, workflow miniaturization, and cost benefits are some of the valuable features of the technology. After the recent advances, NGS emerged as a proven alternative for classical Sanger sequencing in the typing of human leukocyte antigens (HLA). By virtue of the clonal amplification of single DNA molecules ambiguous typing results can be avoided. Simultaneously, a higher sample throughput can be achieved by tagging of DNA molecules with multiplex identifiers and pooling of PCR products before sequencing. In our experience, up to 380 samples can be typed for HLA-A, -B, and -DRB1 in high-resolution during every sequencing run. In molecular oncology, NGS shows a markedly increased sensitivity in comparison to the conventional Sanger sequencing and is developing to the standard diagnostic tool in detection of somatic mutations in cancer cells with great impact on personalized treatment of patients.
Keywords: HLA; Molecular oncology; NGS.
Figures

References
-
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. - PubMed
-
- Glenn TC. Field guide to next-generation DNA sequencers. Mol Ecol Res. 2011;11:759–769. - PubMed
-
- Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–696. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

